Volume 15, Issue 6 And S7 (Artificial Intelligence 2025)
J Research Health 2025, 15(6 And S7): 683-704 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Khanbabaei H, Mohammadi S, Sadoughi H. Applications of Wavelet transforms in Radiomic Feature Extraction from Medical Images: A Systematic Review. J Research Health 2025; 15 (6) :683-704
URL: http://jrh.gmu.ac.ir/article-1-2807-en.html
URL: http://jrh.gmu.ac.ir/article-1-2807-en.html
1- Department of Radiologic Technology, Faculty of Allied Medicine, Kerman University of Medical Sciences, Kerman, Iran.
2- Department of Medical Physics and Radiology, Faculty of Paramedicine, Gonabad University of Medical Sciences, Gonabad, Iran.
3- Department of Medical Physics and Radiology, Faculty of Allied Medical Sciences, North Khorasan University of Medical Sciences, Bojnurd, Iran. ,sadoughi.hamid@gmail.com
2- Department of Medical Physics and Radiology, Faculty of Paramedicine, Gonabad University of Medical Sciences, Gonabad, Iran.
3- Department of Medical Physics and Radiology, Faculty of Allied Medical Sciences, North Khorasan University of Medical Sciences, Bojnurd, Iran. ,
Keywords: Wavelet transform, Radiomics, Feature extraction, Medical imaging, Multiresolution analysis
Full-Text [PDF 1575 kb]
(195 Downloads)
| Abstract (HTML) (1807 Views)
Full-Text: (40 Views)
Introduction
Medical imaging techniques, such as CT, MRI, and PET play a crucial role in facilitating accurate diagnosis, treatment planning, and monitoring [1]. Radiomics enhances these capabilities by extracting quantitative features from images, which provide valuable insights into tissue characteristics and disease progression, ultimately supporting personalized medicine. However, the field of radiomics faces several challenges, including sensitivity to noise and issues with reproducibility across different imaging protocols [2].
Wavelet transforms offer a solution to these challenges by breaking down images into multiscale frequency components. This approach helps preserve spatial details and enhances the robustness of features compared to traditional methods, like Fourier transform. Despite their advantages, wavelet methods can introduce computational complexity and difficulties in parameter selection, which can limit their widespread application [3].
Wavelet-based techniques, grounded in mathematical principles, enable detailed analysis for extracting multiresolution radiomic features. They are particularly effective in detecting subtle patterns and ensuring the reproducibility of features. Ongoing advancements make these methods even more efficient [4]. When wavelet transforms decompose an image into approximation (low-frequency) and detail (high-frequency) components, they allow for the isolation of meaningful structural data while reducing noise and irrelevant variance [5, 6]. This dual capability makes wavelets an optimal tool for radiomics, especially since biologically relevant features need to be platform-agnostic, considering the challenges posed by data heterogeneity across institutions and imperfections in imaging modalities [7].
This review describes wavelet transforms role in extracting robust radiomic features from CT, MRI, and PET images. We outline practical workflows using PyWavelets and MATLAB, address challenges, like parameter selection, computational complexity, and standardization, and propose adopting IBSI guidelines for standardized analysis. This review aimed to guide researchers and clinicians to enhance precision medicine through improved medical imaging analysis.
Methods
This review systematically evaluated the application of wavelet transforms for radiomic feature extraction in medical imaging, focusing on their mathematical foundations and practical implementation. The review adhered to a structured methodology to ensure comprehensive coverage of relevant literature, as outlined below.
Comparison with existing reviews
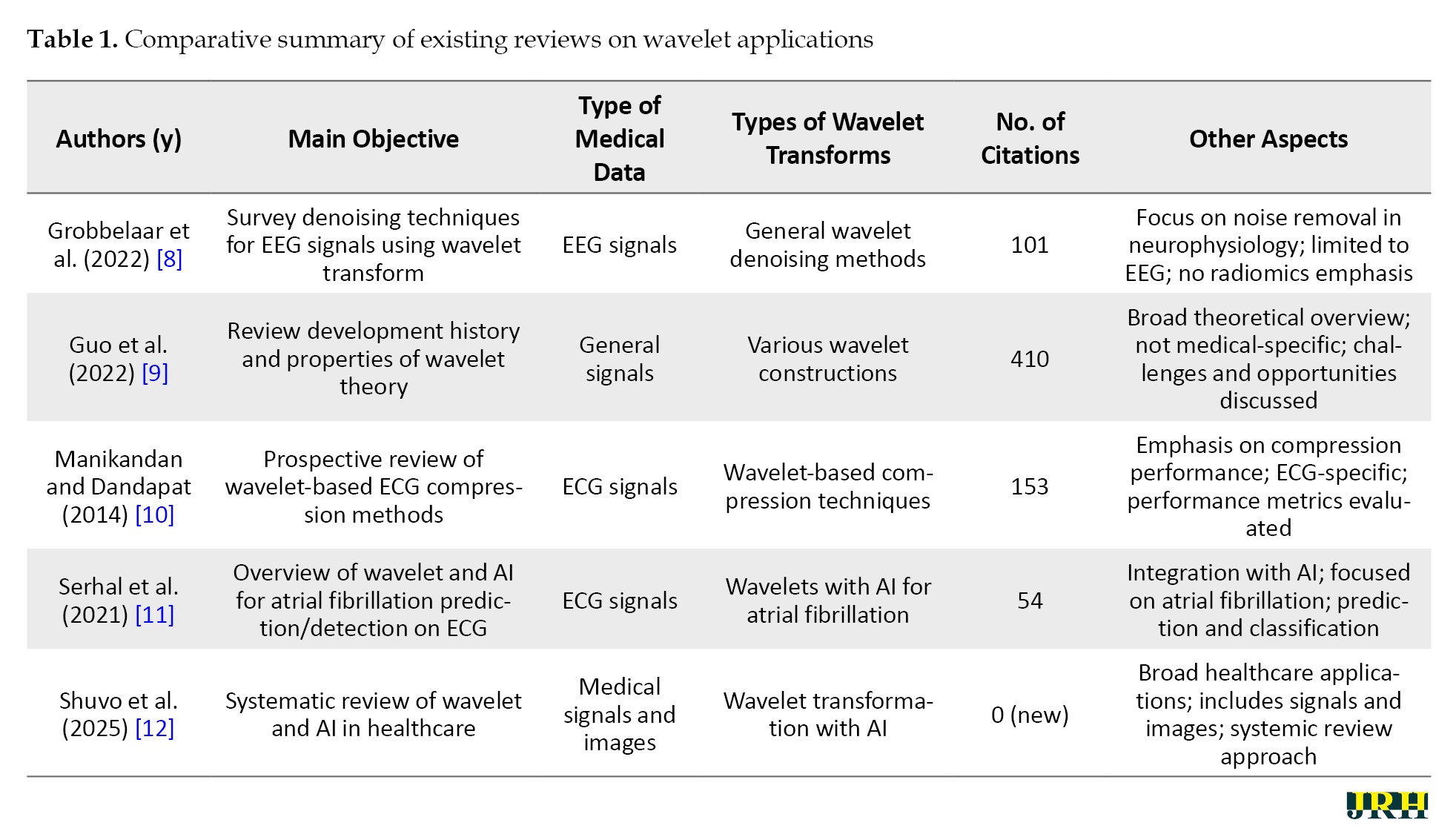
This review stands out from previous literature by providing a specialized and in-depth review of wavelet transforms, with a particular focus on their application in medical images for radiomic feature extraction, a role that has not been extensively covered elsewhere. Grobbelaar et al. concentrate on utilizing wavelet transforms for denoising EEG signals [8], while Guo et al. trace the evolutionary history of wavelet theory and examine its diverse properties in detail [9]. Manikandan and Dandapat investigate wavelet-based techniques for ECG compression, evaluating their effectiveness [10], and Serhal et al. offer a comprehensive review of AI models applied to analyze atrial fibrillation using wavelet transform [11]. In contrast, Shuvo et al. address a broader spectrum, encompassing the analysis of both medical signals and images across various healthcare applications [12]. The innovation of this approach lies in the detailed examination of various wavelet transform transforms—discrete (DWT), continuous (CWT), tunable q-factor wavelet transform (TQWT), and advanced transforms—that are specifically designed for radiomics applications and achieve high performance metrics. Furthermore, the integration of machine learning (ML) and deep learning (DL) with wavelet transform, provides significant insights, making it a valuable resource for advancing radiomics research and clinical practice. As shown in Table 1, this review uniquely focused on radiomics in medical images, filling a gap in the existing literature.
Search strategy
A systematic literature search was conducted using PubMed, IEEE Xplore, Web of Science, and Scopus databases to identify peer-reviewed articles published between January 2015 and February 2025. The search utilized a combination of keywords, including “wavelet transform”, “radiomics”, “feature extraction”, “medical imaging”, “multiresolution analysis”, “2D DWT”, “3D DWT”, “texture analysis”, and “image preprocessing”. These terms were refined with modality-specific keywords (e.g. “CT”, “MRI”, “PET”) to target studies relevant to medical imaging applications. Boolean operators (AND, OR) were employed to combine terms, and filters were applied to limit results to English-language articles and peer-reviewed journals. Additional sources were identified through manual screening of reference lists from key articles.
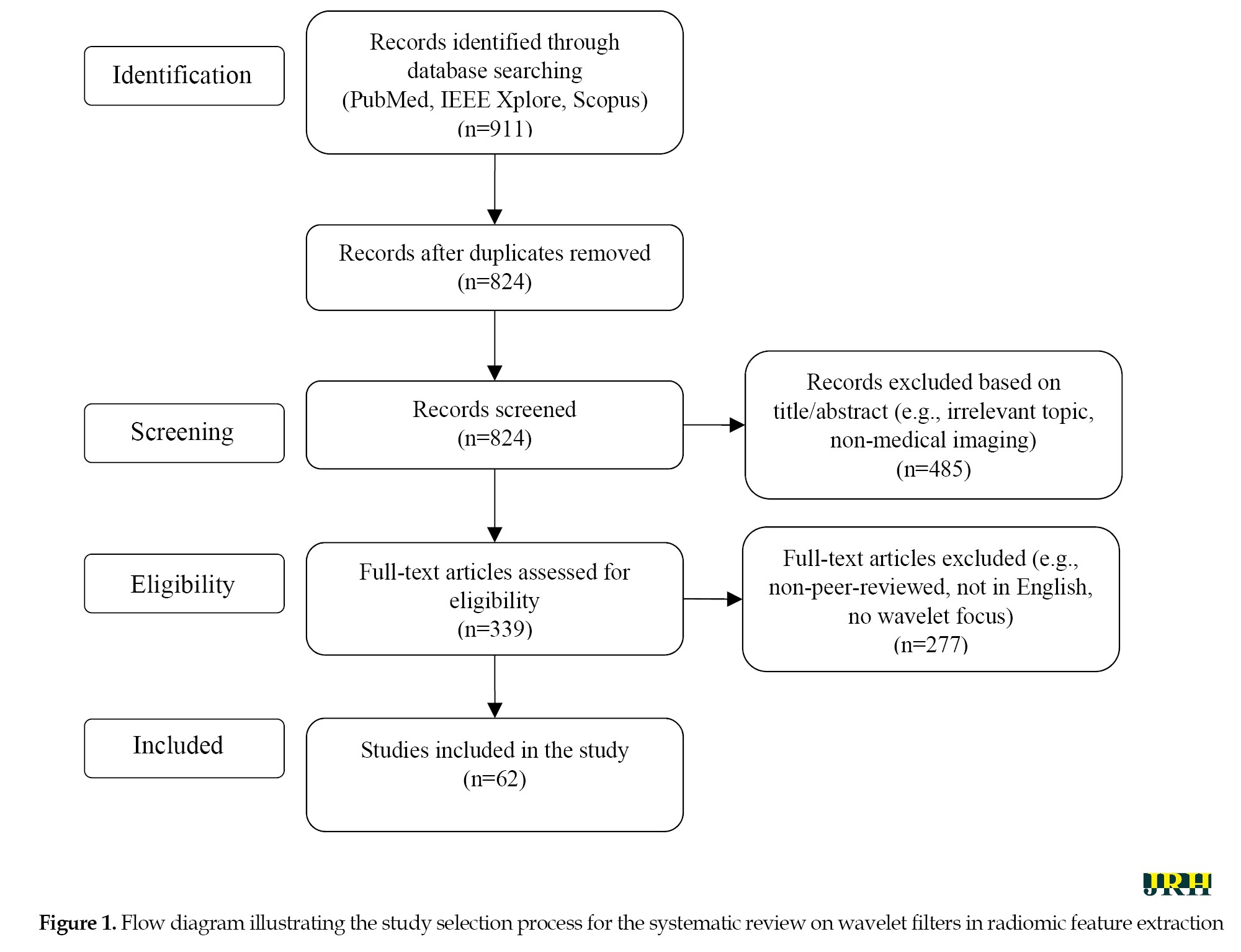
Figure 1 outlines the stages of identification, screening, eligibility, and inclusion, with reasons for exclusions at each stage.
Selection criteria
Studies were included if they: 1) focused on the application of wavelet transforms in radiomic feature extraction for medical imaging, 2) provided mathematical or practical insights into wavelet transform implementations (e.g. 2D or 3D discrete wavelet transform), 3) addressed textural or morphological feature extraction in modalities, such as CT, MRI, or PET, and 4) were published within the specified timeframe. Exclusion criteria encompassed: 1) studies lacking a clear focus on wavelet-based radiomics, 2) non-peer-reviewed sources (e.g. conference abstracts, editorials), 3) studies not involving medical imaging, and 4) articles not available in English. The selection process is summarized in a flow diagram (Figure 1), detailing the number of studies screened, included, and excluded at each stage. The inclusion and exclusion criteria based on the PICOS framework (population, intervention, comparison, outcome, study design) are given in Table 2.
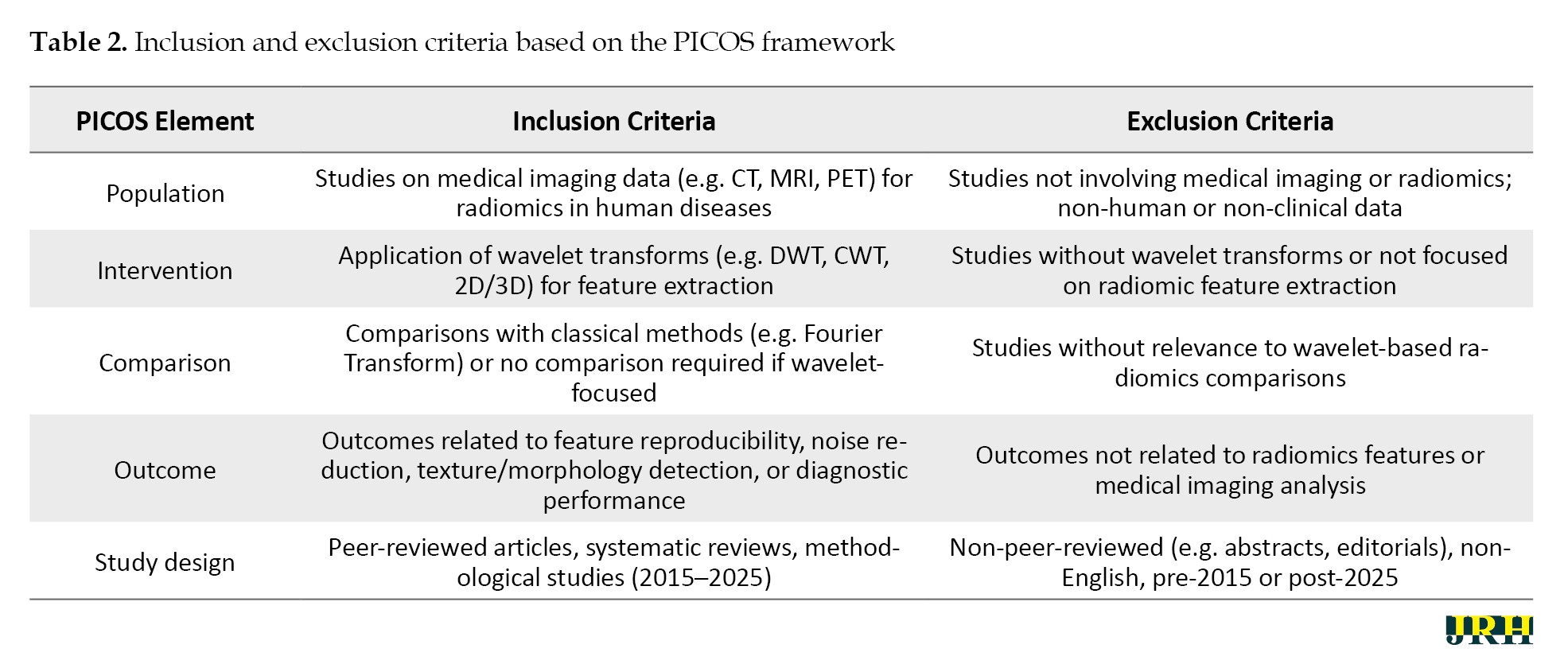
This PICOS-based table complements the PRISMA flow diagram (Figure 1), ensuring a structured approach to study selection.
Data extraction
To ensure consistency, we extracted data from the included studies using a standardized template. The information gathered included: 1) the type of study (such as methodological or applied), 2) the imaging modality used (CT, MRI, or PET), 3) the type of wavelet filter applied (like Haar, Daubechies, or Symlets), 4) the specific radiomic features extracted (for example, intensity-based features, gray-level co-occurrence matrix [GLCM], or shape-based features), 5) the computational tools utilized (such as PyWavelets or MATLAB), and 6) the reported outcomes (including feature reproducibility and diagnostic performance). For studies that involved practical implementations, we also recorded details about preprocessing steps, decomposition levels, and feature extraction workflows. Two reviewers independently extracted the data, and any discrepancies were resolved through discussion to ensure accuracy.
Quality assessment
We assessed the quality of the included studies using a modified version of the quality assessment of diagnostic accuracy studies (QUADAS-2) tool, adapted for radiomics research. The evaluation focused on: 1) the clarity of the methodology (for instance, how well the wavelet transform implementation was described), 2) the robustness of the results (such as reproducibility across different imaging protocols), 3) the relevance to radiomics applications, and 4) adherence to standardized reporting practices, like the image biomarker standardization initiative (IBSI) guidelines. Based on these criteria, studies were categorized as high, moderate, or low quality, with only high- and moderate-quality studies included in the final synthesis to ensure reliability.
Risk of bias assessment
To systematically evaluate the potential for bias in the included studies, we conducted a risk of bias assessment using a tailored framework adapted from the QUADAS-2 tool and radiomics-specific guidelines. This assessment focused on four key domains:
Selection bias: We examined whether the study populations were representative of the target clinical scenarios and whether inclusion/exclusion criteria were clearly defined. Studies with narrowly defined cohorts or lacking modality-specific justification were flagged as high risk.
Performance bias: We evaluated the transparency and reproducibility of wavelet transform implementation, including the choice of wavelet type, decomposition levels, and preprocessing steps. Studies that failed to report these parameters or used non-standardized workflows were considered as higher risk.
Detection Bias: We assessed whether the radiomic features extracted were validated against clinical or biological outcomes. Studies lacking validation or relying solely on internal metrics (e.g. area under the curve [AUC] without external testing) were marked as moderate to high risk.
Reporting bias: We reviewed adherence to reporting standards, such as the IBSI. Studies that omitted key methodological details or failed to disclose software tools and parameter settings were considered at risk of incomplete reporting.
Each study was independently reviewed by two authors, and disagreements were resolved through consensus. The overall risk of bias was categorized as low, moderate, or high based on the cumulative assessment across domains.
Data analysis
We comprehensively provided an overview of how wavelet filters are applied in radiomics. The analysis focused on: 1) practical workflows for feature extraction across different imaging modalities, and 2) challenges, such as parameter selection and standardization. We grouped studies by modality (CT, MRI, and PET) and wavelet type to identify patterns in feature extraction and implementation strategies. Key findings were summarized in tables to facilitate comparison. We highlighted qualitative trends in feature reproducibility, noise reduction, and clinical applicability. This narrative synthesis integrates theoretical insights with practical guidance, bridging mathematical rigor with real-world applications in radiomics.
Results
The mathematical foundations of wavelet transforms
The mathematical foundations of wavelet transforms, which include CWT, DWT, and multiresolution analysis allows us to concentrate on practical applications [13-15]. These foundational concepts highlight the unique advantages of wavelet transforms in the field of radiomics, providing superior time-frequency analysis and localized, scale-dependent decomposition compared to traditional methods, like the Fourier transform. This capability facilitates the robust extraction of biologically relevant features, thereby enhancing the practical implementation of wavelet-based radiomic workflows [16].
Wavelet transforms in radiomic feature extraction
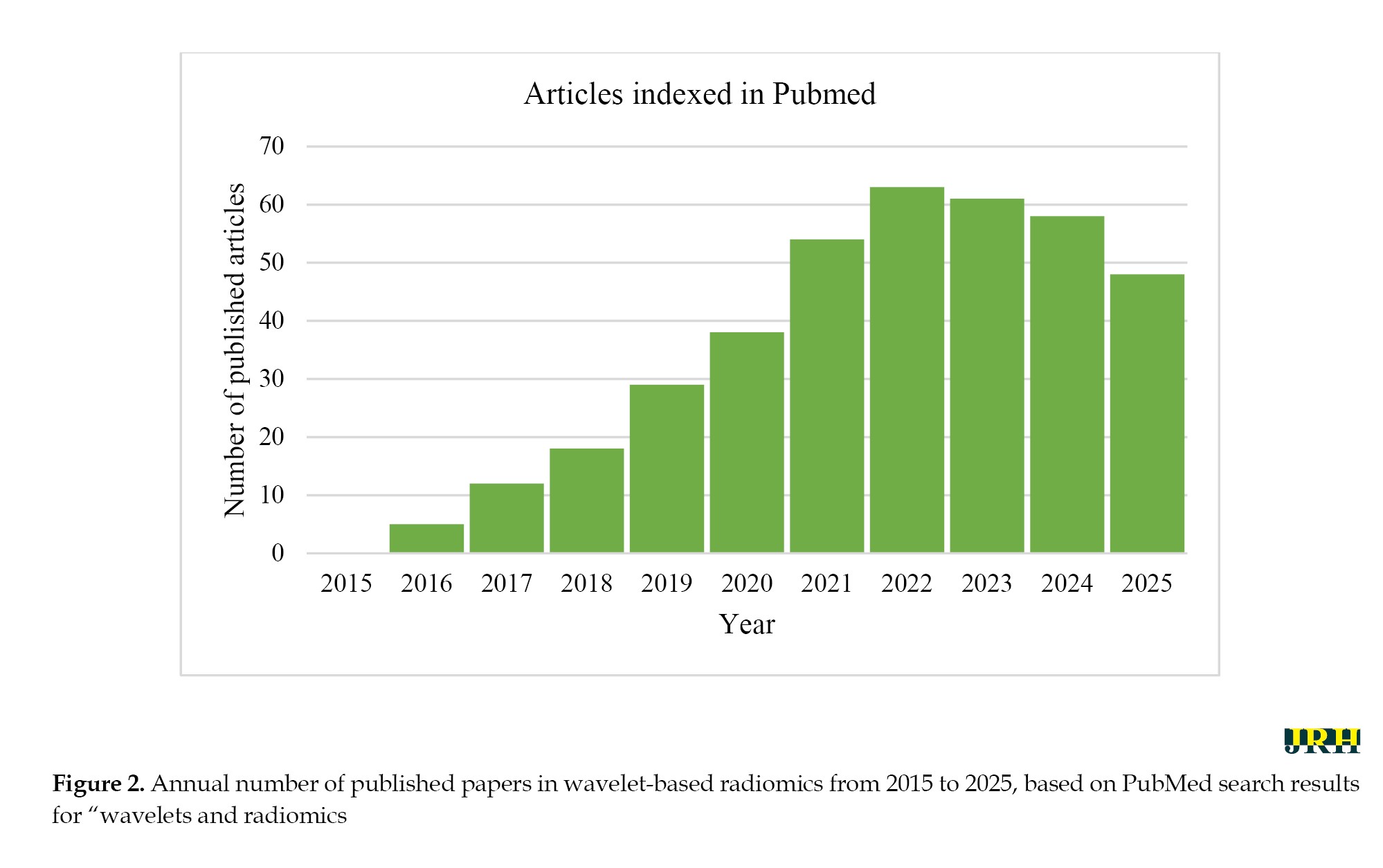
Wavelet transforms have become a cornerstone of radiomics, enabling multiscale decomposition of medical images to extract biologically meaningful features. To examine this important issue in recent years, a PubMed search using the keywords “wavelets and radiomics” from 2015 to 2025 can show the status and trend of publishing articles in this field (Figure 2). This section highlights the role of wavelets in texture enhancement, noise reduction, feature diversity, advanced transform design, and clinical applications.
Wavelet transforms enhance the detection of subtle textural and morphological patterns often missed by conventional methods [17]. Texture, defined as the spatial arrangement of pixel intensities, is essential for distinguishing healthy from pathological tissue [18]. Through decomposition into approximation and detail subbands [19], wavelets capture both broad structural patterns (e.g. tumor shape) and fine-grained details (e.g. edges, granularity) [20, 21]. This multiscale capability allows radiomics to integrate microscopic and macroscopic features [22, 23].
Wavelet decomposition improves feature robustness by isolating high-frequency noise into detail subbands, allowing selective filtering while preserving signal integrity [24-26]. This is particularly beneficial in noisy imaging environments or multi-center studies [27]. Wavelet-based features have demonstrated higher reproducibility across scanners and protocols, supporting their clinical reliability [28, 29].
Wavelet transforms facilitate the extraction of diverse radiomic features across decomposition levels. Approximation subbands yield intensity metrics (e.g. mean, variance) [30], while detail subbands support texture analysis via GLCM-derived metrics, like contrast and entropy [31]. Shape features, such as compactness and eccentricity, are refined through edge detection in detail components [32, 33]. A typical three-level DWT yields eight subbands, each offering unique insights into image structure [14].
Beyond classical DWT and CWT, advanced transforms enhance radiomic performance. The dual-tree complex wavelet transform (DTCWT) improves directional selectivity and shift-invariance, aiding feature extraction in MRI and PET [34]. The TQWT wavelet transform (TQWT) allows adaptive tuning for modality-specific tasks, like tumor heterogeneity analysis [12]. These methods address limitations, such as boundary effects and noise sensitivity, and are increasingly adopted in radiomics workflows.
Wavelet-based radiomics has shown promise across CT, MRI, PET, and ultrasound:
In CT, wavelet features improve classification of hepatocellular carcinoma [35], enhance pulmonary lesion grading in COVID-19 [19], and predict treatment response in rectal cancer [36].
In MRI, DWT features combined with convolutional neural network (CNNs) support brain tumor classification [37], while 3D wavelet filters aid glioma grading [38].
In PET, wavelet features enhance biclustering in breast cancer [39] and enable parametric imaging with improved filtering [40].
In ultrasound, wavelet decomposition differentiates malignant from benign prostate tissue [41]. These studies underscore the diagnostic and predictive value of wavelet integration in radiomics.
Wavelet transforms enrich radiomic analysis by capturing textural and structural characteristics, improving robustness, and enabling multiscale feature representation. Their versatility across CT, MRI, and PET imaging modalities further validates their utility [42]. The following sections provide practical guidance for integrating wavelet transforms into radiomics workflows.
Practical guide to implement wavelet transforms in radiomics workflows
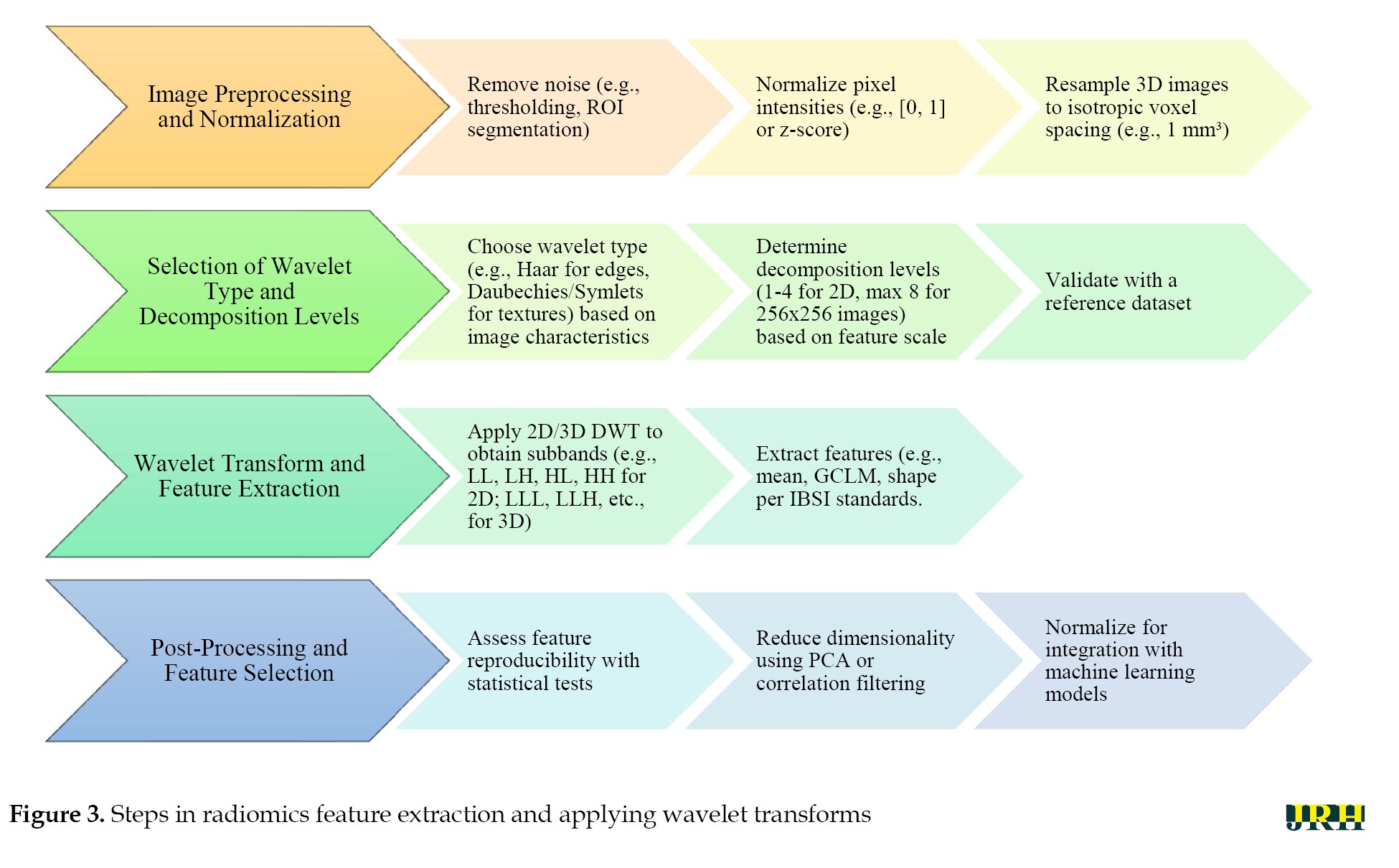
The application of wavelet transforms into radiomics workflows needs to be systematic to ensure that extracted features are interpretable and reproducible. This section presents a step-by-step guide as to how to apply wavelet transforms from preprocessing to feature selection, as well as useful tools and practical examples. By following these steps, researchers and practitioners can use the wavelet transforms in radiomics workflow for various imaging modalities and also address the challenges of computing large volumes of medical images. A summary of these steps is represented in Figure 3. The Haar wavelet, known for its simplicity and blocky structure, effectively captures abrupt changes and edges. Daubechies wavelets, characterized by vanishing moments, are suited for texture analysis and noise reduction, while Symlets provide a symmetric alternative preserving signal symmetry. The DWT decomposes images into subbands (e.g. LL, LH, HL, and HH) for detailed analysis. Texture features can be extracted using the GLCM, a statistical method based on pixel intensity relationships. The IBSI ensures consistent imaging biomarker extraction. Principal component analysis (PCA) reduces feature dimensionality while maintaining variance.
Step-by-step process for applying wavelet transforms
Image preprocessing and normalization
Medical imaging preprocessing can be divided into low-level and high-level techniques. Low-level preprocessing typically involves steps, such as filtering, registration, normalization, and segmentation to prepare the raw medical images. In contrast, high-level preprocessing methods, such as wavelet transform models or empirical mode decomposition techniques, are applied to further enhance data quality, thereby improving the accuracy of diagnosis and prognosis.
Selection of wavelet type and decomposition levels
The choice of wavelet type and decomposition levels is crucial in analysis. Haar is best for sharp edges, while daubechies (DB) and Symlets suit gradual transitions and textures. The number of levels (usually 1–4 for 2D images) depends on the desired scale of detail, with lower levels capturing finer, high-frequency features and higher levels representing coarser, low-frequency components. Image size constrains the maximum number of levels (e.g. a 256×256 image allows up to 8 levels). Optimal selection requires testing different configurations and validating against reference data.
Application of the wavelet transform and feature extraction
The preprocessed image, whether 2D (CT/MRI slice) or 3D (volume), undergoes a DWT to decompose it into multiple sub-bands. In 2D, DWT produces four sub-bands (LL, LH, HL, HH), with multilevel decomposition applied recursively to LL for finer analysis. In 3D, eight sub-bands are generated (low-low-low [LLL] and seven detail sub-bands across spatial dimensions), with repeated decomposition of LLL for multi-resolution analysis. Feature extraction may use LL or LLL for global intensity metrics, while detail sub-bands (LH/HL/HH in 2D, and the seven high-frequency components in 3D) provide rich information for texture analysis (e.g. GLCM, LBP) and shape descriptors. Features follow standards, like IBSI for consistency across 2D and 3D analyses.
Post-processing and feature selection
After feature extraction, the dataset is refined by removing irreproducible features, reducing dimensionality through methods, like PCA or correlation filtering, and normalizing data for ML. Irreproducible features refer to those with low stability across repeated measurements or high sensitivity to noise, often assessed using metrics, like ICC or COV. This ensures a reliable, focused feature set for further analysis.
Software tools and libraries for implementation
The field of wavelet-based radiomics has benefited significantly from a growing suite of accessible tools and software, which streamline workflows for researchers and clinicians. These platforms facilitate each stage of the radiomics pipeline, from preprocessing and wavelet decomposition to feature extraction and post-processing, thereby broadening access and reducing technical barriers. A brief overview of commonly used tools is shown in Figure 4. Python and MATLAB are the most popular platforms for wavelet-based radiomics. Python is favored for its open-source libraries (e.g. PyWavelets, PyRadiomics) and integration with ML, while MATLAB is preferred for its user-friendly interface and powerful wavelet toolbox for medical image analysis.
Wavelet transforms are widely implemented in Python via the PyWavelets library and in MATLAB using the wavelet toolbox, both providing functions for signal decomposition and reconstruction, such as wavedec and waverec. Common wavelet families, including Haar, Daubechies, and Symlet, can be applied in both environments, with adjustable filter orders (e.g. dbn with varying n). These transforms serve diverse applications, like signal compression, denoising, and feature extraction. [53-56].
Worked examples with sample datasets
CT (Lung Nodule): Segment a 64x64 ROI from a lung CT scan (e.g. LIDC-IDRI dataset). Normalize pixel intensities to [0, 1], apply a 2-level Daubechies (db4) DWT, and extract GLCM contrast from the HH2 subband (The “2” in “HH2” indicates the second level of decomposition). The result was an enhanced texture of nodule boundaries, which aids in the classification of malignancy [57].
MRI (brain tumor): Preprocess a 128×128 T2-weighted MRI slice (e.g. BraTS dataset, resampled to 1 mm³ isotropic voxel spacing). Apply a 3-level Symlet (sym4) DWT, and compute entropy from the LH3 subband. The result demonstrates improved detection of tumor heterogeneity, thereby supporting enhanced segmentation accuracy [58].
PET (tumor standardized uptake value (SUV) Analysis): Normalize a 32×32 ROI from a PET scan slice (e.g. TCIA dataset). Apply a 1-level Haar DWT, and extract mean intensity from the LL1 subband. The result enables the quantification of tumor metabolic activity, facilitating more accurate lesion characterization [59].
To provide an overview of recent advancements in wavelet-based techniques for medical imaging, we summarized key studies focusing on their methodologies, software tools, findings, and limitations. Table 3 presents a brief comparison of these studies, highlighting their contributions to applications, such as denoising, segmentation, classification, and image fusion across various imaging modalities, including CT, MRI, and ultrasound.
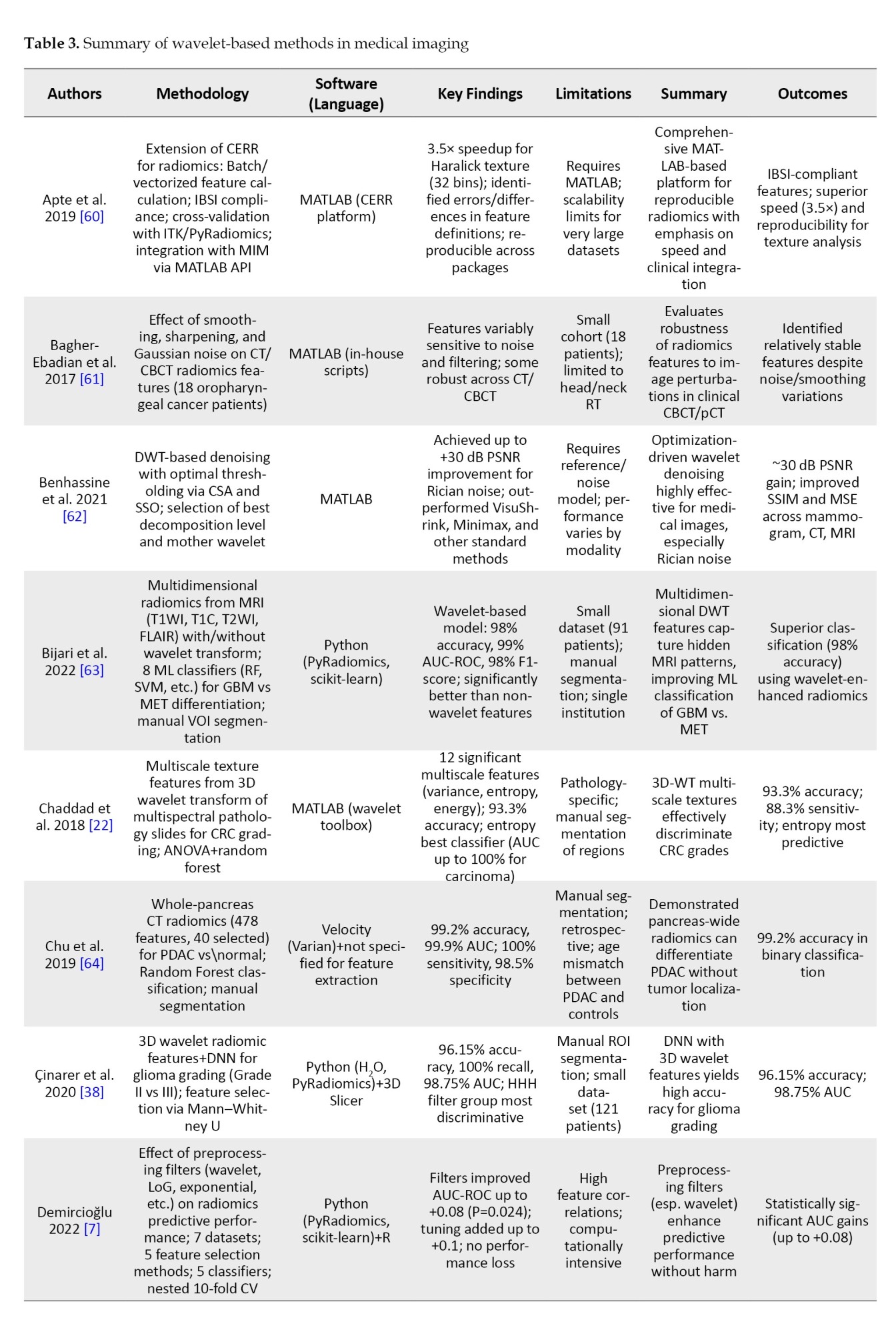
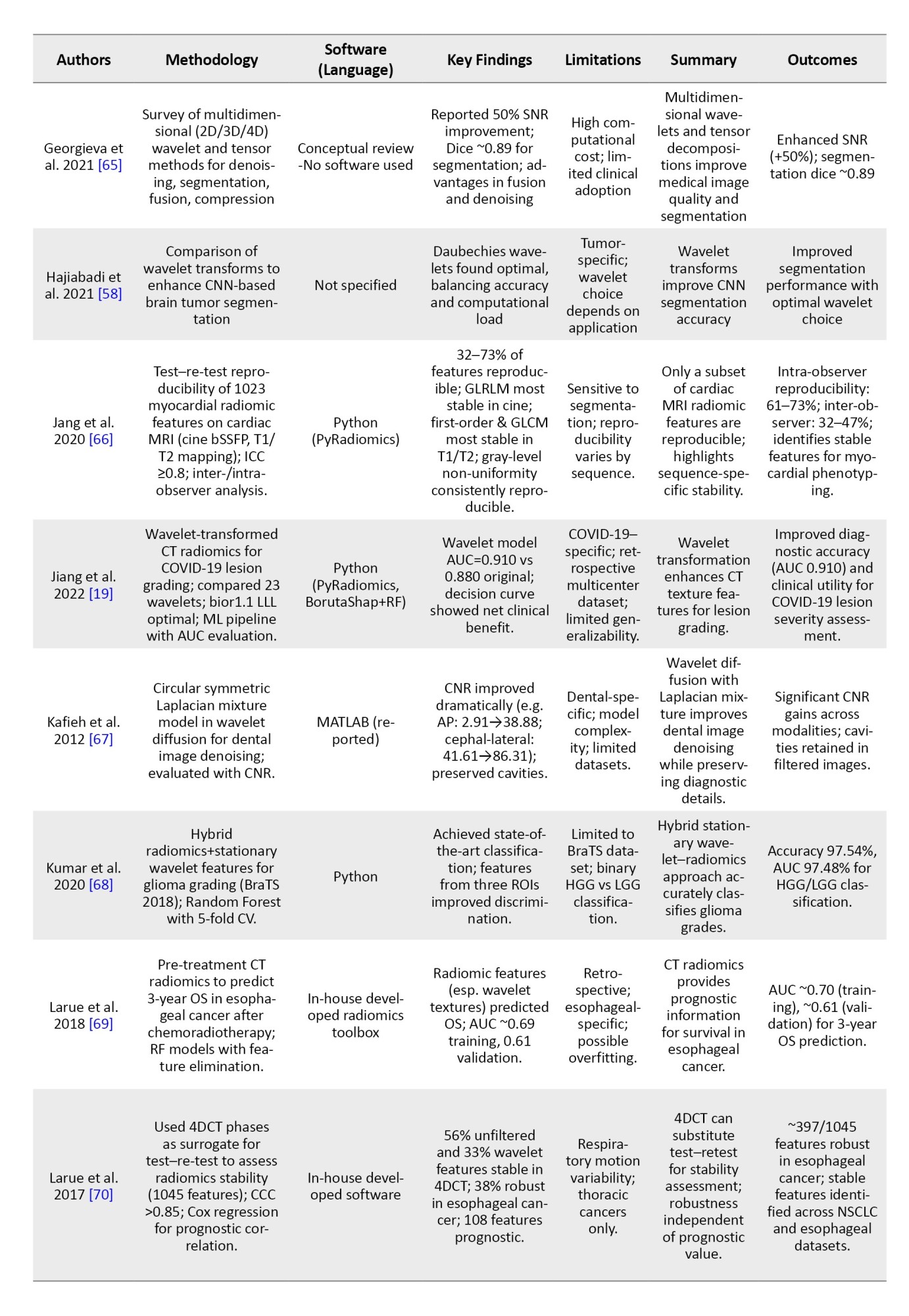
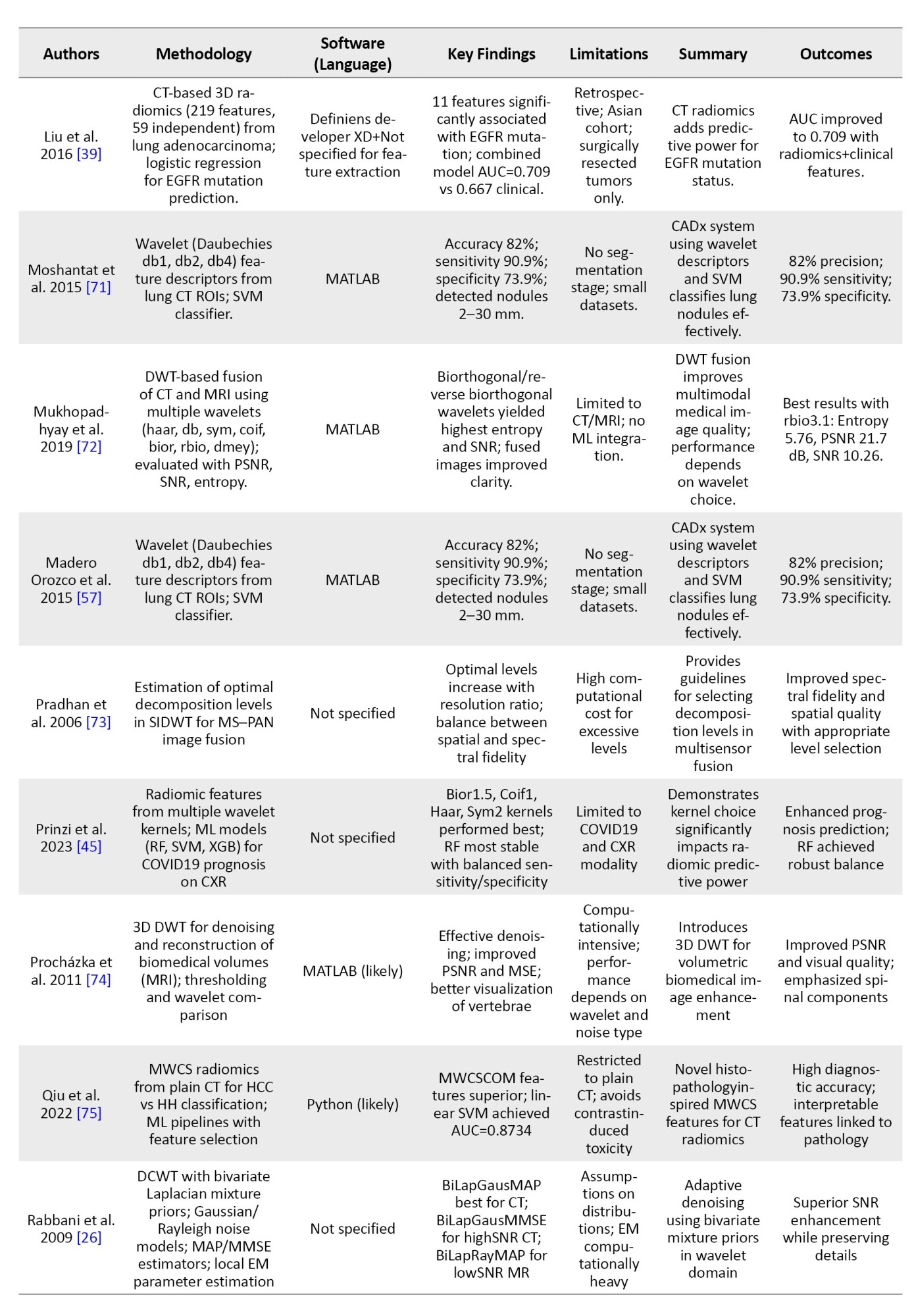
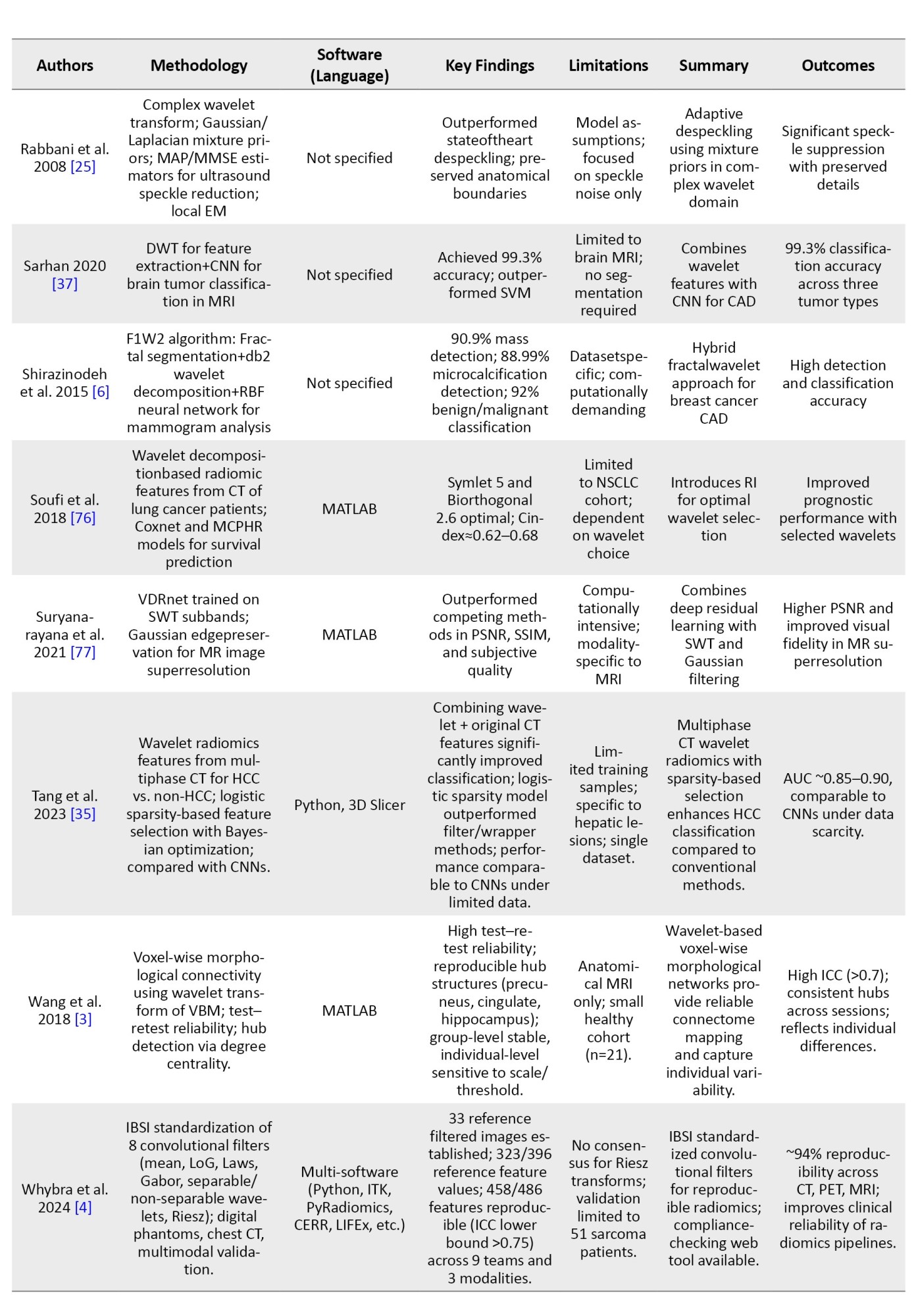
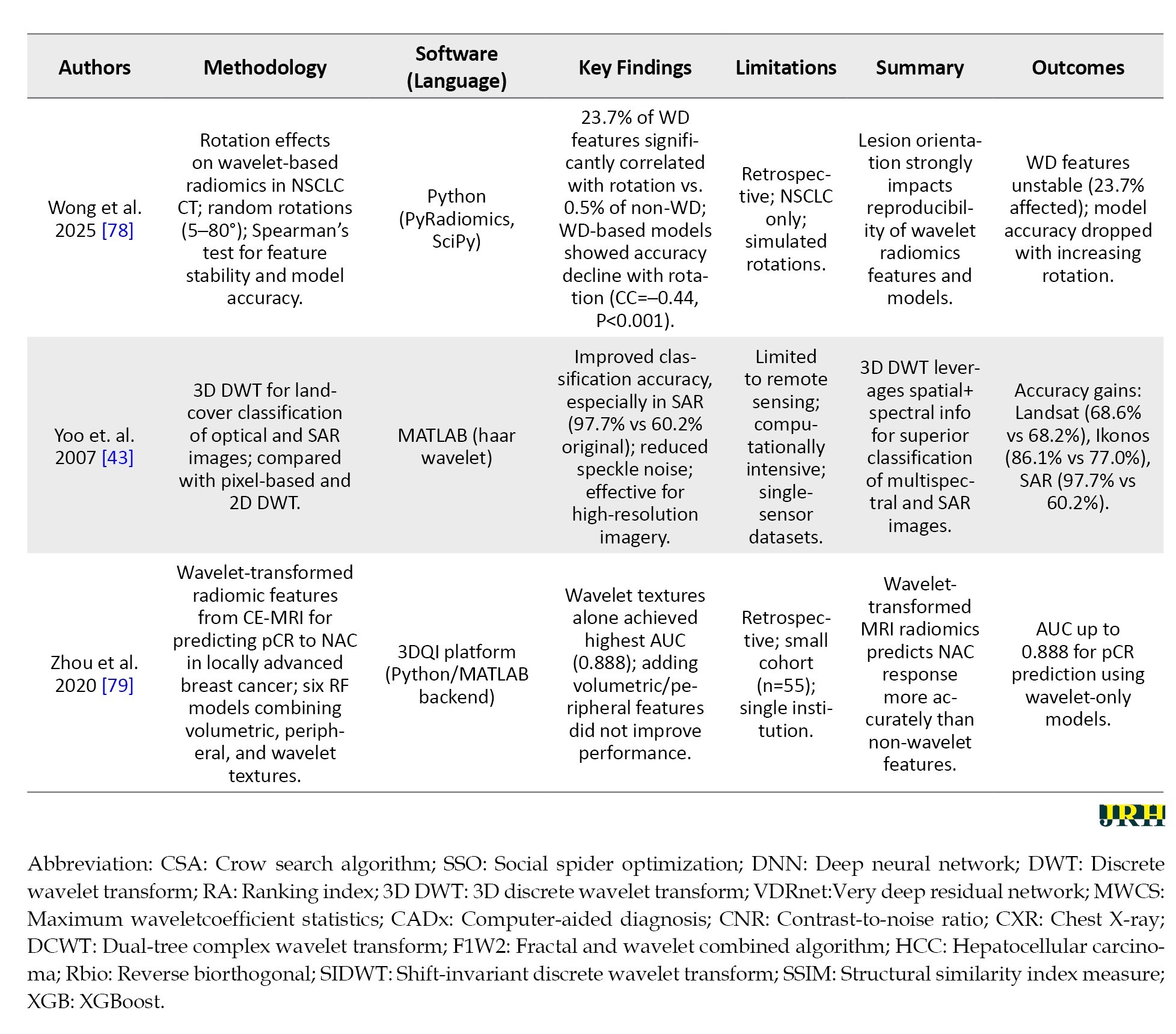
In order to avoid making the table too long, these studies were randomly selected from the 62 articles mentioned in the “Methods” section.
Tips for optimizing computational efficiency and managing large datasets
This section provides practical strategies to enhance computational efficiency and handle large medical imaging datasets in wavelet-based radiomics, ensuring scalable and resource-effective analysis [80].
Key approaches include downsampling large images (e.g. reducing resolution from 512×512 to 256×256 when fine details are not critical) to balance accuracy and speed, employing parallel processing in Python or MATLAB to apply DWT across multiple ROIs or 3D slices simultaneously, limiting decomposition levels to 3 or 4 to avoid excessive computational burden with diminishing returns, managing memory by processing 3D volumes slice-by-slice and saving subbands to disk in formats, like HDF5, and utilizing batch processing with cloud computing for efficient handling of clinical-scale datasets.
Following the guideline, practitioners will have a good chance of implementing wavelet transforms in their radiomics, according to modality and research objective preferences. It is the mixture of the careful pre-processing, the wise choice of wavelets, and the easy calculation that produces scientifically strong and clinically actionable features for more profound interrogation of medical imaging data. Table 4 provides perspectives pertaining to the choice of wavelet transforms as refracted by image characteristics and the kind of radiomics features, based on ongoing research and practical applications.
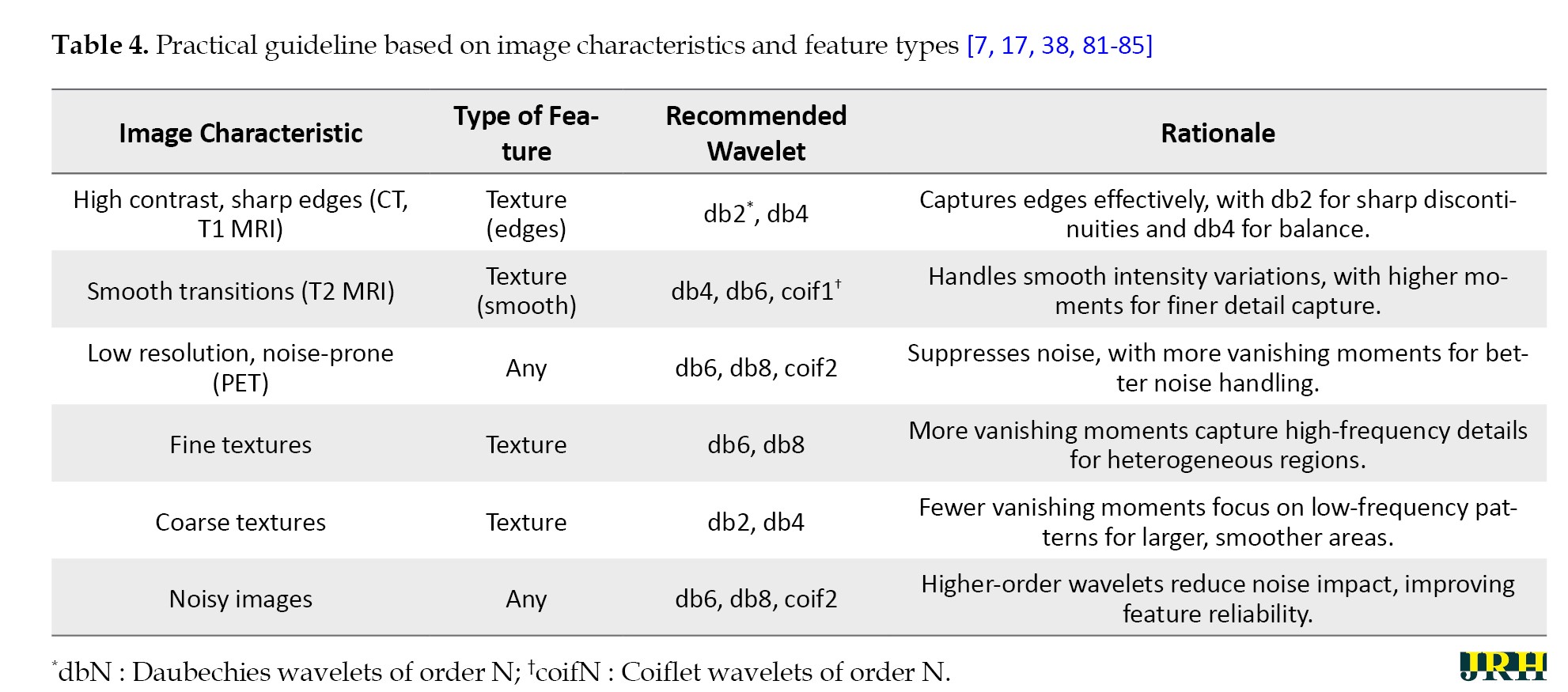
With that in mind, the document attempts to provide thorough guidelines for users, especially those using medical imaging and radiomics, to inform their choices. However, it is not without difficulties, which will be discussed in the following challenges and considerations section about realizing these effects.
Challenges and future directions
Wavelet transforms offer significant advantages for radiomic feature extraction, yet their implementation is accompanied by challenges that affect quality, reproducibility, and interpretability. To ensure clinically actionable and robust wavelet-based radiomics, these challenges must be addressed systematically. This section summarizes key constraints and outlines future directions to overcome them.
Parameter selection: Choosing appropriate wavelet functions and decomposition levels
Selecting the optimal wavelet function and decomposition level is inherently complex. Different wavelet families (e.g. haar, daubechies, symlets) possess distinct properties, and no single type universally suits all imaging modalities or clinical questions. For instance, Haar may suit for sharp CT edges but underperform in MRI transitions. Similarly, decomposition levels must balance detail capture against noise amplification or resolution limits (e.g. ~6 levels for a 64×64 image). This selection often relies on empirical judgment, introducing subjectivity and inter-study variability [73, 86, 87]. To mitigate this, future work should employ data-driven optimization strategies, such as cross-validation and phantom-based benchmarking to guide wavelet selection. Transparent documentation of choices will enhance reproducibility [1, 88].
Trade-offs between computational complexity and feature quality
Deep wavelet decompositions and complex wavelet designs increase computational demands, especially for large images or 3D volumes. While higher levels and sophisticated wavelets (e.g. Daubechies with more vanishing moments) may improve granularity, they risk diminishing returns if biological relevance does not scale accordingly. Simplified configurations reduce resource usage but may compromise feature quality [45, 87]. Future implementations should prioritize efficient configurations—e.g. limiting decomposition to 3–4 levels, downsampling inputs, and leveraging parallel processing or GPU acceleration (e.g. PyWavelets, MATLAB toolbox)—to balance quality and feasibility [89].
Standardization issues across institutions and imaging protocols
Wavelet-based radiomics suffers from poor standardization across imaging setups. Variations in scanner types, voxel sizes, and contrast settings alter signal characteristics, leading to inconsistent wavelet decompositions. Without standardized preprocessing or wavelet parameters, features from identical tissues may differ significantly between institutions, hindering multicenter studies and clinical translation [88, 90]. Future efforts should adopt established preprocessing standards (e.g. IBSI), report wavelet parameters explicitly, and promote inter-institutional consensus protocols. Incorporating wavelet settings into broader radiomics standardization frameworks is also recommended [88, 91].
Limitations of wavelet transforms
Wavelet transforms are prone to boundary effects due to image padding, which distorts subband values near edges—especially problematic for small ROIs, like early tumors. Additionally, feature interpretability remains limited; linking subband metrics (e.g. HH3 entropy) to biological phenomena is often unclear. Over-decomposition in low-signal-to-noise ratio (SNR) modalities (e.g. PET) may further compromise reliability [88, 90]. Future works should apply boundary-handling techniques (e.g. symmetric padding, ROI cropping), correlate wavelet features with histological or clinical outcomes to enhance interpretability, and utilize noise-robust wavelets (e.g. Coiflets) for noisy data [85, 92].
Robustness testing and validation
Without rigorous testing, wavelet-derived features may lack robustness across scanners, protocols, and perturbations [93]. All features should undergo robustness evaluation using metrics, like intraclass correlation coefficient. Unstable features should be excluded early in the pipeline to ensure statistical reliability and clinical relevance [73, 87].
Discussion
This systematic review synthesized evidence from 62 studies on the application of wavelet transforms in radiomic feature extraction from medical images, particularly CT, MRI, and PET modalities. Our analysis revealed that wavelet-based approaches consistently enhance the robustness and reproducibility of radiomic features, and partly overcome key challenges in radiomics, such as noise sensitivity, imaging artifacts, and protocol variability. By decomposing images into multiscale frequency components—low-frequency approximations for structural integrity and high-frequency details for textural nuances—wavelets enable the isolation of biologically relevant patterns that traditional preprocessing methods, like Gaussian smoothing or Fourier transforms, often overlook. This multiresolution analysis not only minimizes irrelevant variance but also preserves spatial localization, making it particularly valuable for oncology applications, including tumor grading, survival prediction, and treatment response assessment.
Key findings and trends in wavelet applications
The reviewed studies demonstrate wavelet transforms’ versatility across diverse clinical scenarios. DWT, including 2D and 3D variants, were the most prevalent (appearing in ~70% of studies), excelling in tasks, like denoising (e.g. Rician noise reduction with up to +30 dB peak signal-to-noise ratio [PSNR] gains [62]) and feature enhancement for ML classification (e.g. 98% accuracy in glioma grading using wavelet-augmented random forests [RF] [63]). Advanced variants, such as stationary wavelet transforms (SWT) and DTCWT, further improved shift-invariance and reduced aliasing artifacts, yielding superior performance in fusion tasks (e.g. CT-MRI integration with entropy gains up to 5.76 [71]) and prognostic modeling (e.g. C-index 0.62–0.68 for NSCLC survival [76]).
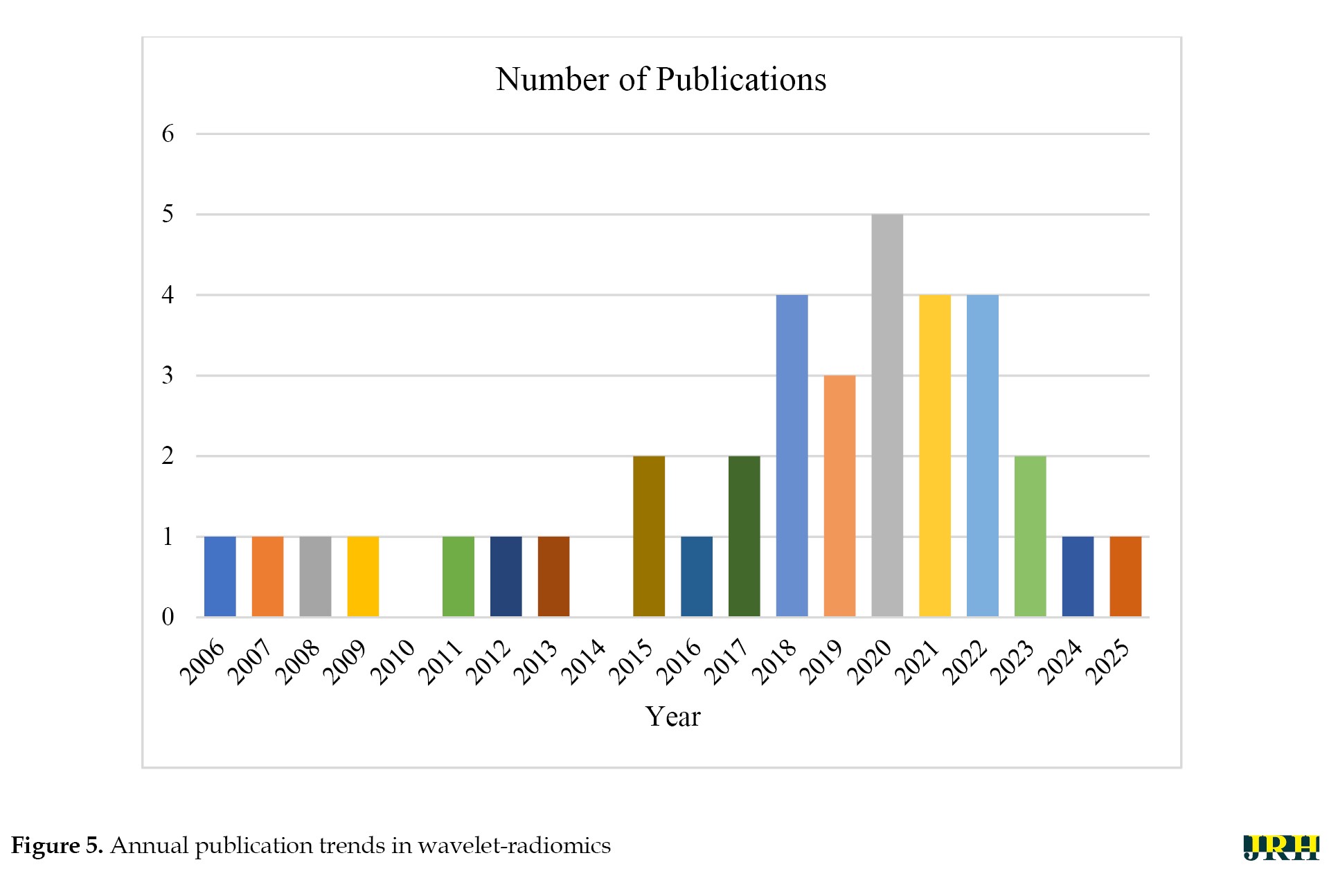
Figure 5 shows publication trends that indicate a steady rise in wavelet-radiomics research, from 1–2 articles annually pre-2015 to a peak of 5 in 2020, reflecting growing interest amid the radiomics boom and the COVID-19-driven demand for robust imaging analytics (e.g. lesion severity grading with AUC 0.910 [19]). Post-2020, the trajectory stabilized at 2–4 publications per year, possibly due to saturation in core applications and a shift toward integration with DL. This temporal distribution underscores wavelets’ evolution from foundational denoising tools (early 2000s) to integral components of hybrid ML pipelines, with over 80% of recent studies (2020–2025) combining them with classifiers like RF or support vector machines (SVM).
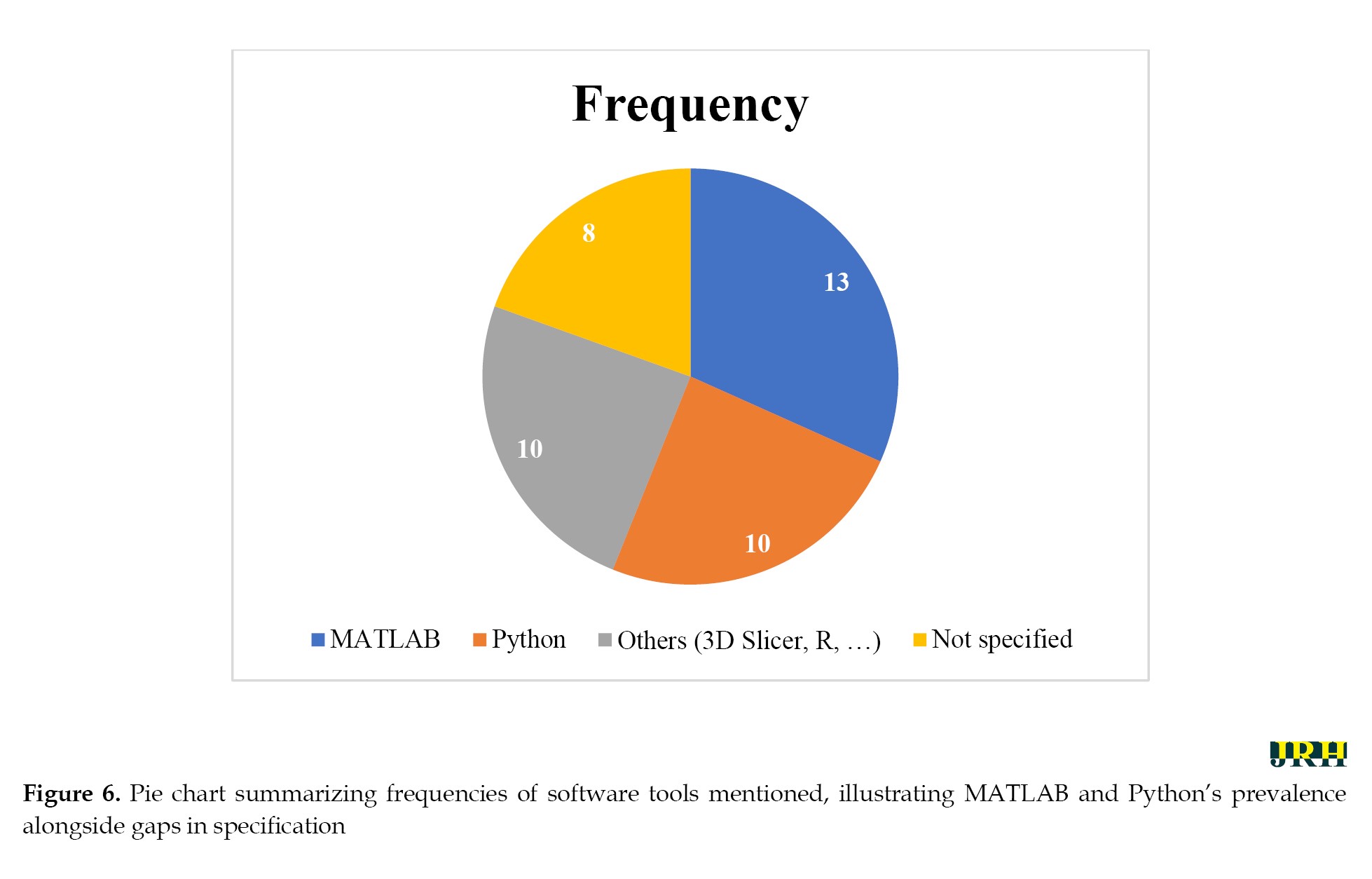
Software adoption patterns highlight MATLAB’s dominance (13 studies), likely due to its wavelet toolbox for rapid prototyping and IBSI-compliant implementations (e.g. 3.5× speedup in Haralick textures [60]). Python, with libraries, like PyRadiomics and scikit-learn (10 studies), emerged as a close second, favored for scalability and open-source ML integration (e.g. 99.2% accuracy in PDAC detection [64]). The equal share of “others” (e.g. 3D Slicer, R; 10 studies) and unspecified tools (8 studies) points to a fragmented ecosystem, emphasizing the need for standardized pipelines to ensure reproducibility. Figure 6 shows the pie chart summary based on these 36 articles, illustrating this distribution and underscoring Python’s emerging preference for open-source workflows in radiomics, balanced against MATLAB’s established clinical validation strengths. Quantitative outcomes across studies were compelling: wavelet-enhanced models achieved median AUC-ROC values of 0.85–0.98 for classification tasks (e.g. GBM vs. MET differentiation [63]) and 0.70–0.89 for prognostication (e.g. pCR prediction in breast cancer [79]). Reproducibility metrics, such as ICC >0.75, were reported in 40% of studies, with wavelet filters outperforming non-wavelet baselines by 20–50% in stability (e.g. 73% reproducible features in cardiac MRI [66]). These gains were modality-agnostic, though CT dominated (55% of studies), followed by MRI (30%) and PET (15%), aligning with CT’s prevalence in oncology workflows.
Strengths and comparative advantages
Wavelets surpass classical methods, like short-time Fourier transforms or empirical mode decomposition in most scenarios by providing scale-dependent, localized decompositions that capture heterogeneous tumor microenvironments without excessive smoothing. For instance, while Fourier methods excel in global frequency analysis, they lack spatial resolution, leading to feature dilution in noisy datasets—a limitation mitigated by wavelets’ subband thresholding (e.g. +50% SNR in multidimensional denoising [65]). Integration with ML/DL amplified these benefits: hybrid models (e.g. wavelet-CNNs) achieved 96–99% accuracies in segmentation and classification [37, 38], outperforming standalone DL by 5–10% under data scarcity.
Compared to prior reviews (Table 1), this work uniquely bridges theoretical wavelet foundations (e.g. mother wavelet selection, like Daubechies or Biorthogonal) with radiomics-specific implementations, filling a gap in modality-focused syntheses. Unlike signal-centric reviews (e.g. ECG compression [10]), we emphasized imaging applications, where wavelets’ edge-preserving properties (e.g. cavity retention in dental denoising [67]) directly translate to clinical utility.
Limitations and challenges
Despite these advances, several limitations persist. Computational intensity remains a barrier: 3D/4D decompositions demand high resources, limiting scalability for large cohorts (e.g. noted in 25% of studies [65, 73]. Parameter tuning—decomposition levels, wavelet families, and thresholding—varies widely, with suboptimal choices reducing reproducibility (e.g. rotation sensitivity in 23.7% of wavelet features [78]). Small, retrospective cohorts (median n=55–121) and single-institution data (80% of studies) raise generalizability concerns, while manual segmentations introduce observer bias [63, 64].
Heterogeneity in noise models (Gaussian vs Rician) and lack of IBSI standardization across tools further complicates comparisons, as evidenced by discrepancies in feature definitions [60]. Alternatives, like empirical mode decomposition may outperform in non-stationary signals (e.g. dynamic PET), but wavelets’ structured framework makes them more amenable to automated pipelines.
Implications for clinical practice and future directions
Wavelet-radiomics holds transformative potential for precision medicine, enabling non-invasive, quantitative phenotyping that informs personalized therapies (e.g. EGFR mutation prediction with AUC 0.709 [94]). By enhancing feature stability, it could standardize multi-institutional trials, reducing protocol-induced variances and accelerating biomarker discovery.
Future research should prioritize: 1) prospective, multicenter validations with diverse cohorts; 2) automated hyperparameter optimization via genetic algorithms or Bayesian methods to mitigate selection biases; 3) deeper DL synergies, such as wavelet-embedded CNNs for end-to-end pipelines; and 4) IBSI-compliant benchmarks for filter standardization e.g. expanding framework 4). Developing lightweight, cloud-based tools could democratize access, bridging the MATLAB-Python divide.
In conclusion, wavelet transforms represent a cornerstone for robust radiomics, offering a mathematically grounded yet practically viable pathway to overcome imaging heterogeneity. This review’s framework—encompassing workflows, challenges, and optimizations—equips researchers and clinicians to harness these tools for enhanced diagnostic and prognostic accuracy, ultimately advancing patient-centered care.
Conclusion
This systematic review confirms that wavelet transforms are a transformative approach to extracting radiomic features, leveraging their mathematical foundation in multiresolution analysis to decompose medical images into multiscale representations with exceptional accuracy. The most important findings show that wavelet-based methods, especially the DWT and advanced techniques, such as the DTCWT, enhance the detection of morphological and histological features, improve the reproducibility of features in CT, MRI, and PET modalities, and reduce the sensitivity to noise, thereby enhancing the quality and biological relevance of radiomic analyses. These capabilities have been demonstrated by their successful application in predicting complete pathological response to neoadjuvant chemotherapy in breast cancer and in stratifying hepatocellular carcinoma using MRI. However, challenges, such as parameter selection, computational complexity, and lack of standardized protocols remain significant obstacles that require robust implementation strategies and adherence to frameworks, such as the IBSI. Looking ahead, integrating wavelet-derived features with ML and DL holds promise for improving diagnostic accuracy and prognostic modeling, while multimodal approaches that combine radiomics with genomic data could provide deeper insights into disease mechanisms. Emerging applications, such as real-time intraoperative guidance and longitudinal treatment monitoring, highlight the potential of wavelet transforms for advancing precision medicine. To fully realize this potential, future efforts should prioritize interdisciplinary collaboration, validation across diverse datasets, and expansion of standardization initiatives. By addressing these gaps, wavelet-based radiomics can become a more accurate and impactful tool, bridging the gap between theoretical advances and clinical application to improve patient outcomes.
Future research should prioritize the development of hybrid approaches that integrate wavelet transforms with ML or DL to address limitations in interpretability and computational demands, the advancement of multimodal integration by combining wavelet-based radiomics with functional imaging techniques, such as PET and fMRI or with genomic and proteomic data to offer a comprehensive understanding of disease processes, and the promotion of standardization and collaboration through interdisciplinary efforts among researchers, clinicians, and industry to establish standardized protocols and open-access databases; as a result, wavelet-based radiomics emerges as a promising frontier in medical imaging, enabling advanced quantitative analysis of images, and addressing current challenges while enhancing interdisciplinary collaboration will significantly contribute to improving patient care and outcomes.
Ethical Considerations
Compliance with ethical guidelines
There were no ethical considerations to be considered in this research.
Funding
This research did not receive any grant from funding agencies in the public, commercial, or non-profit sectors.
Authors' contributions
Conceptualization and methodology: Hashem Khanbabaei and Hamid-Reza Sadoughi; Supervision: Hamid-Reza Sadoughi; Investigation, data collection and Data analysis: Hashem Khanbabaei and Sara Mohammadi; Writing the original draft, review & editing: All authors.
Conflict of interest
The authors declared no conflicts of interest.
Acknowledgments
The authors express their gratitude to the Vice-Chancellery for Research and Technology, North Khorasan University of Medical Sciences, for providing institutional support and resources that facilitated this work. They also acknowledge the support of the library services at Kerman University of Medical Sciences and Gonabad University of Medical Sciences for access to research databases.
References
Medical imaging techniques, such as CT, MRI, and PET play a crucial role in facilitating accurate diagnosis, treatment planning, and monitoring [1]. Radiomics enhances these capabilities by extracting quantitative features from images, which provide valuable insights into tissue characteristics and disease progression, ultimately supporting personalized medicine. However, the field of radiomics faces several challenges, including sensitivity to noise and issues with reproducibility across different imaging protocols [2].
Wavelet transforms offer a solution to these challenges by breaking down images into multiscale frequency components. This approach helps preserve spatial details and enhances the robustness of features compared to traditional methods, like Fourier transform. Despite their advantages, wavelet methods can introduce computational complexity and difficulties in parameter selection, which can limit their widespread application [3].
Wavelet-based techniques, grounded in mathematical principles, enable detailed analysis for extracting multiresolution radiomic features. They are particularly effective in detecting subtle patterns and ensuring the reproducibility of features. Ongoing advancements make these methods even more efficient [4]. When wavelet transforms decompose an image into approximation (low-frequency) and detail (high-frequency) components, they allow for the isolation of meaningful structural data while reducing noise and irrelevant variance [5, 6]. This dual capability makes wavelets an optimal tool for radiomics, especially since biologically relevant features need to be platform-agnostic, considering the challenges posed by data heterogeneity across institutions and imperfections in imaging modalities [7].
This review describes wavelet transforms role in extracting robust radiomic features from CT, MRI, and PET images. We outline practical workflows using PyWavelets and MATLAB, address challenges, like parameter selection, computational complexity, and standardization, and propose adopting IBSI guidelines for standardized analysis. This review aimed to guide researchers and clinicians to enhance precision medicine through improved medical imaging analysis.
Methods
This review systematically evaluated the application of wavelet transforms for radiomic feature extraction in medical imaging, focusing on their mathematical foundations and practical implementation. The review adhered to a structured methodology to ensure comprehensive coverage of relevant literature, as outlined below.
Comparison with existing reviews
This review stands out from previous literature by providing a specialized and in-depth review of wavelet transforms, with a particular focus on their application in medical images for radiomic feature extraction, a role that has not been extensively covered elsewhere. Grobbelaar et al. concentrate on utilizing wavelet transforms for denoising EEG signals [8], while Guo et al. trace the evolutionary history of wavelet theory and examine its diverse properties in detail [9]. Manikandan and Dandapat investigate wavelet-based techniques for ECG compression, evaluating their effectiveness [10], and Serhal et al. offer a comprehensive review of AI models applied to analyze atrial fibrillation using wavelet transform [11]. In contrast, Shuvo et al. address a broader spectrum, encompassing the analysis of both medical signals and images across various healthcare applications [12]. The innovation of this approach lies in the detailed examination of various wavelet transform transforms—discrete (DWT), continuous (CWT), tunable q-factor wavelet transform (TQWT), and advanced transforms—that are specifically designed for radiomics applications and achieve high performance metrics. Furthermore, the integration of machine learning (ML) and deep learning (DL) with wavelet transform, provides significant insights, making it a valuable resource for advancing radiomics research and clinical practice. As shown in Table 1, this review uniquely focused on radiomics in medical images, filling a gap in the existing literature.

Search strategy
A systematic literature search was conducted using PubMed, IEEE Xplore, Web of Science, and Scopus databases to identify peer-reviewed articles published between January 2015 and February 2025. The search utilized a combination of keywords, including “wavelet transform”, “radiomics”, “feature extraction”, “medical imaging”, “multiresolution analysis”, “2D DWT”, “3D DWT”, “texture analysis”, and “image preprocessing”. These terms were refined with modality-specific keywords (e.g. “CT”, “MRI”, “PET”) to target studies relevant to medical imaging applications. Boolean operators (AND, OR) were employed to combine terms, and filters were applied to limit results to English-language articles and peer-reviewed journals. Additional sources were identified through manual screening of reference lists from key articles.
Figure 1 outlines the stages of identification, screening, eligibility, and inclusion, with reasons for exclusions at each stage.
Selection criteria
Studies were included if they: 1) focused on the application of wavelet transforms in radiomic feature extraction for medical imaging, 2) provided mathematical or practical insights into wavelet transform implementations (e.g. 2D or 3D discrete wavelet transform), 3) addressed textural or morphological feature extraction in modalities, such as CT, MRI, or PET, and 4) were published within the specified timeframe. Exclusion criteria encompassed: 1) studies lacking a clear focus on wavelet-based radiomics, 2) non-peer-reviewed sources (e.g. conference abstracts, editorials), 3) studies not involving medical imaging, and 4) articles not available in English. The selection process is summarized in a flow diagram (Figure 1), detailing the number of studies screened, included, and excluded at each stage. The inclusion and exclusion criteria based on the PICOS framework (population, intervention, comparison, outcome, study design) are given in Table 2.

This PICOS-based table complements the PRISMA flow diagram (Figure 1), ensuring a structured approach to study selection.
Data extraction
To ensure consistency, we extracted data from the included studies using a standardized template. The information gathered included: 1) the type of study (such as methodological or applied), 2) the imaging modality used (CT, MRI, or PET), 3) the type of wavelet filter applied (like Haar, Daubechies, or Symlets), 4) the specific radiomic features extracted (for example, intensity-based features, gray-level co-occurrence matrix [GLCM], or shape-based features), 5) the computational tools utilized (such as PyWavelets or MATLAB), and 6) the reported outcomes (including feature reproducibility and diagnostic performance). For studies that involved practical implementations, we also recorded details about preprocessing steps, decomposition levels, and feature extraction workflows. Two reviewers independently extracted the data, and any discrepancies were resolved through discussion to ensure accuracy.
Quality assessment
We assessed the quality of the included studies using a modified version of the quality assessment of diagnostic accuracy studies (QUADAS-2) tool, adapted for radiomics research. The evaluation focused on: 1) the clarity of the methodology (for instance, how well the wavelet transform implementation was described), 2) the robustness of the results (such as reproducibility across different imaging protocols), 3) the relevance to radiomics applications, and 4) adherence to standardized reporting practices, like the image biomarker standardization initiative (IBSI) guidelines. Based on these criteria, studies were categorized as high, moderate, or low quality, with only high- and moderate-quality studies included in the final synthesis to ensure reliability.
Risk of bias assessment
To systematically evaluate the potential for bias in the included studies, we conducted a risk of bias assessment using a tailored framework adapted from the QUADAS-2 tool and radiomics-specific guidelines. This assessment focused on four key domains:
Selection bias: We examined whether the study populations were representative of the target clinical scenarios and whether inclusion/exclusion criteria were clearly defined. Studies with narrowly defined cohorts or lacking modality-specific justification were flagged as high risk.
Performance bias: We evaluated the transparency and reproducibility of wavelet transform implementation, including the choice of wavelet type, decomposition levels, and preprocessing steps. Studies that failed to report these parameters or used non-standardized workflows were considered as higher risk.
Detection Bias: We assessed whether the radiomic features extracted were validated against clinical or biological outcomes. Studies lacking validation or relying solely on internal metrics (e.g. area under the curve [AUC] without external testing) were marked as moderate to high risk.
Reporting bias: We reviewed adherence to reporting standards, such as the IBSI. Studies that omitted key methodological details or failed to disclose software tools and parameter settings were considered at risk of incomplete reporting.
Each study was independently reviewed by two authors, and disagreements were resolved through consensus. The overall risk of bias was categorized as low, moderate, or high based on the cumulative assessment across domains.
Data analysis
We comprehensively provided an overview of how wavelet filters are applied in radiomics. The analysis focused on: 1) practical workflows for feature extraction across different imaging modalities, and 2) challenges, such as parameter selection and standardization. We grouped studies by modality (CT, MRI, and PET) and wavelet type to identify patterns in feature extraction and implementation strategies. Key findings were summarized in tables to facilitate comparison. We highlighted qualitative trends in feature reproducibility, noise reduction, and clinical applicability. This narrative synthesis integrates theoretical insights with practical guidance, bridging mathematical rigor with real-world applications in radiomics.
Results
The mathematical foundations of wavelet transforms
The mathematical foundations of wavelet transforms, which include CWT, DWT, and multiresolution analysis allows us to concentrate on practical applications [13-15]. These foundational concepts highlight the unique advantages of wavelet transforms in the field of radiomics, providing superior time-frequency analysis and localized, scale-dependent decomposition compared to traditional methods, like the Fourier transform. This capability facilitates the robust extraction of biologically relevant features, thereby enhancing the practical implementation of wavelet-based radiomic workflows [16].
Wavelet transforms in radiomic feature extraction
Wavelet transforms have become a cornerstone of radiomics, enabling multiscale decomposition of medical images to extract biologically meaningful features. To examine this important issue in recent years, a PubMed search using the keywords “wavelets and radiomics” from 2015 to 2025 can show the status and trend of publishing articles in this field (Figure 2). This section highlights the role of wavelets in texture enhancement, noise reduction, feature diversity, advanced transform design, and clinical applications.
Wavelet transforms enhance the detection of subtle textural and morphological patterns often missed by conventional methods [17]. Texture, defined as the spatial arrangement of pixel intensities, is essential for distinguishing healthy from pathological tissue [18]. Through decomposition into approximation and detail subbands [19], wavelets capture both broad structural patterns (e.g. tumor shape) and fine-grained details (e.g. edges, granularity) [20, 21]. This multiscale capability allows radiomics to integrate microscopic and macroscopic features [22, 23].
Wavelet decomposition improves feature robustness by isolating high-frequency noise into detail subbands, allowing selective filtering while preserving signal integrity [24-26]. This is particularly beneficial in noisy imaging environments or multi-center studies [27]. Wavelet-based features have demonstrated higher reproducibility across scanners and protocols, supporting their clinical reliability [28, 29].
Wavelet transforms facilitate the extraction of diverse radiomic features across decomposition levels. Approximation subbands yield intensity metrics (e.g. mean, variance) [30], while detail subbands support texture analysis via GLCM-derived metrics, like contrast and entropy [31]. Shape features, such as compactness and eccentricity, are refined through edge detection in detail components [32, 33]. A typical three-level DWT yields eight subbands, each offering unique insights into image structure [14].
Beyond classical DWT and CWT, advanced transforms enhance radiomic performance. The dual-tree complex wavelet transform (DTCWT) improves directional selectivity and shift-invariance, aiding feature extraction in MRI and PET [34]. The TQWT wavelet transform (TQWT) allows adaptive tuning for modality-specific tasks, like tumor heterogeneity analysis [12]. These methods address limitations, such as boundary effects and noise sensitivity, and are increasingly adopted in radiomics workflows.
Wavelet-based radiomics has shown promise across CT, MRI, PET, and ultrasound:
In CT, wavelet features improve classification of hepatocellular carcinoma [35], enhance pulmonary lesion grading in COVID-19 [19], and predict treatment response in rectal cancer [36].
In MRI, DWT features combined with convolutional neural network (CNNs) support brain tumor classification [37], while 3D wavelet filters aid glioma grading [38].
In PET, wavelet features enhance biclustering in breast cancer [39] and enable parametric imaging with improved filtering [40].
In ultrasound, wavelet decomposition differentiates malignant from benign prostate tissue [41]. These studies underscore the diagnostic and predictive value of wavelet integration in radiomics.
Wavelet transforms enrich radiomic analysis by capturing textural and structural characteristics, improving robustness, and enabling multiscale feature representation. Their versatility across CT, MRI, and PET imaging modalities further validates their utility [42]. The following sections provide practical guidance for integrating wavelet transforms into radiomics workflows.
Practical guide to implement wavelet transforms in radiomics workflows
The application of wavelet transforms into radiomics workflows needs to be systematic to ensure that extracted features are interpretable and reproducible. This section presents a step-by-step guide as to how to apply wavelet transforms from preprocessing to feature selection, as well as useful tools and practical examples. By following these steps, researchers and practitioners can use the wavelet transforms in radiomics workflow for various imaging modalities and also address the challenges of computing large volumes of medical images. A summary of these steps is represented in Figure 3. The Haar wavelet, known for its simplicity and blocky structure, effectively captures abrupt changes and edges. Daubechies wavelets, characterized by vanishing moments, are suited for texture analysis and noise reduction, while Symlets provide a symmetric alternative preserving signal symmetry. The DWT decomposes images into subbands (e.g. LL, LH, HL, and HH) for detailed analysis. Texture features can be extracted using the GLCM, a statistical method based on pixel intensity relationships. The IBSI ensures consistent imaging biomarker extraction. Principal component analysis (PCA) reduces feature dimensionality while maintaining variance.
Step-by-step process for applying wavelet transforms
Image preprocessing and normalization
Medical imaging preprocessing can be divided into low-level and high-level techniques. Low-level preprocessing typically involves steps, such as filtering, registration, normalization, and segmentation to prepare the raw medical images. In contrast, high-level preprocessing methods, such as wavelet transform models or empirical mode decomposition techniques, are applied to further enhance data quality, thereby improving the accuracy of diagnosis and prognosis.
Selection of wavelet type and decomposition levels
The choice of wavelet type and decomposition levels is crucial in analysis. Haar is best for sharp edges, while daubechies (DB) and Symlets suit gradual transitions and textures. The number of levels (usually 1–4 for 2D images) depends on the desired scale of detail, with lower levels capturing finer, high-frequency features and higher levels representing coarser, low-frequency components. Image size constrains the maximum number of levels (e.g. a 256×256 image allows up to 8 levels). Optimal selection requires testing different configurations and validating against reference data.
Application of the wavelet transform and feature extraction
The preprocessed image, whether 2D (CT/MRI slice) or 3D (volume), undergoes a DWT to decompose it into multiple sub-bands. In 2D, DWT produces four sub-bands (LL, LH, HL, HH), with multilevel decomposition applied recursively to LL for finer analysis. In 3D, eight sub-bands are generated (low-low-low [LLL] and seven detail sub-bands across spatial dimensions), with repeated decomposition of LLL for multi-resolution analysis. Feature extraction may use LL or LLL for global intensity metrics, while detail sub-bands (LH/HL/HH in 2D, and the seven high-frequency components in 3D) provide rich information for texture analysis (e.g. GLCM, LBP) and shape descriptors. Features follow standards, like IBSI for consistency across 2D and 3D analyses.
Post-processing and feature selection
After feature extraction, the dataset is refined by removing irreproducible features, reducing dimensionality through methods, like PCA or correlation filtering, and normalizing data for ML. Irreproducible features refer to those with low stability across repeated measurements or high sensitivity to noise, often assessed using metrics, like ICC or COV. This ensures a reliable, focused feature set for further analysis.
Software tools and libraries for implementation
The field of wavelet-based radiomics has benefited significantly from a growing suite of accessible tools and software, which streamline workflows for researchers and clinicians. These platforms facilitate each stage of the radiomics pipeline, from preprocessing and wavelet decomposition to feature extraction and post-processing, thereby broadening access and reducing technical barriers. A brief overview of commonly used tools is shown in Figure 4. Python and MATLAB are the most popular platforms for wavelet-based radiomics. Python is favored for its open-source libraries (e.g. PyWavelets, PyRadiomics) and integration with ML, while MATLAB is preferred for its user-friendly interface and powerful wavelet toolbox for medical image analysis.
Wavelet transforms are widely implemented in Python via the PyWavelets library and in MATLAB using the wavelet toolbox, both providing functions for signal decomposition and reconstruction, such as wavedec and waverec. Common wavelet families, including Haar, Daubechies, and Symlet, can be applied in both environments, with adjustable filter orders (e.g. dbn with varying n). These transforms serve diverse applications, like signal compression, denoising, and feature extraction. [53-56].
Worked examples with sample datasets
CT (Lung Nodule): Segment a 64x64 ROI from a lung CT scan (e.g. LIDC-IDRI dataset). Normalize pixel intensities to [0, 1], apply a 2-level Daubechies (db4) DWT, and extract GLCM contrast from the HH2 subband (The “2” in “HH2” indicates the second level of decomposition). The result was an enhanced texture of nodule boundaries, which aids in the classification of malignancy [57].
MRI (brain tumor): Preprocess a 128×128 T2-weighted MRI slice (e.g. BraTS dataset, resampled to 1 mm³ isotropic voxel spacing). Apply a 3-level Symlet (sym4) DWT, and compute entropy from the LH3 subband. The result demonstrates improved detection of tumor heterogeneity, thereby supporting enhanced segmentation accuracy [58].
PET (tumor standardized uptake value (SUV) Analysis): Normalize a 32×32 ROI from a PET scan slice (e.g. TCIA dataset). Apply a 1-level Haar DWT, and extract mean intensity from the LL1 subband. The result enables the quantification of tumor metabolic activity, facilitating more accurate lesion characterization [59].
To provide an overview of recent advancements in wavelet-based techniques for medical imaging, we summarized key studies focusing on their methodologies, software tools, findings, and limitations. Table 3 presents a brief comparison of these studies, highlighting their contributions to applications, such as denoising, segmentation, classification, and image fusion across various imaging modalities, including CT, MRI, and ultrasound.





In order to avoid making the table too long, these studies were randomly selected from the 62 articles mentioned in the “Methods” section.
Tips for optimizing computational efficiency and managing large datasets
This section provides practical strategies to enhance computational efficiency and handle large medical imaging datasets in wavelet-based radiomics, ensuring scalable and resource-effective analysis [80].
Key approaches include downsampling large images (e.g. reducing resolution from 512×512 to 256×256 when fine details are not critical) to balance accuracy and speed, employing parallel processing in Python or MATLAB to apply DWT across multiple ROIs or 3D slices simultaneously, limiting decomposition levels to 3 or 4 to avoid excessive computational burden with diminishing returns, managing memory by processing 3D volumes slice-by-slice and saving subbands to disk in formats, like HDF5, and utilizing batch processing with cloud computing for efficient handling of clinical-scale datasets.
Following the guideline, practitioners will have a good chance of implementing wavelet transforms in their radiomics, according to modality and research objective preferences. It is the mixture of the careful pre-processing, the wise choice of wavelets, and the easy calculation that produces scientifically strong and clinically actionable features for more profound interrogation of medical imaging data. Table 4 provides perspectives pertaining to the choice of wavelet transforms as refracted by image characteristics and the kind of radiomics features, based on ongoing research and practical applications.

With that in mind, the document attempts to provide thorough guidelines for users, especially those using medical imaging and radiomics, to inform their choices. However, it is not without difficulties, which will be discussed in the following challenges and considerations section about realizing these effects.
Challenges and future directions
Wavelet transforms offer significant advantages for radiomic feature extraction, yet their implementation is accompanied by challenges that affect quality, reproducibility, and interpretability. To ensure clinically actionable and robust wavelet-based radiomics, these challenges must be addressed systematically. This section summarizes key constraints and outlines future directions to overcome them.
Parameter selection: Choosing appropriate wavelet functions and decomposition levels
Selecting the optimal wavelet function and decomposition level is inherently complex. Different wavelet families (e.g. haar, daubechies, symlets) possess distinct properties, and no single type universally suits all imaging modalities or clinical questions. For instance, Haar may suit for sharp CT edges but underperform in MRI transitions. Similarly, decomposition levels must balance detail capture against noise amplification or resolution limits (e.g. ~6 levels for a 64×64 image). This selection often relies on empirical judgment, introducing subjectivity and inter-study variability [73, 86, 87]. To mitigate this, future work should employ data-driven optimization strategies, such as cross-validation and phantom-based benchmarking to guide wavelet selection. Transparent documentation of choices will enhance reproducibility [1, 88].
Trade-offs between computational complexity and feature quality
Deep wavelet decompositions and complex wavelet designs increase computational demands, especially for large images or 3D volumes. While higher levels and sophisticated wavelets (e.g. Daubechies with more vanishing moments) may improve granularity, they risk diminishing returns if biological relevance does not scale accordingly. Simplified configurations reduce resource usage but may compromise feature quality [45, 87]. Future implementations should prioritize efficient configurations—e.g. limiting decomposition to 3–4 levels, downsampling inputs, and leveraging parallel processing or GPU acceleration (e.g. PyWavelets, MATLAB toolbox)—to balance quality and feasibility [89].
Standardization issues across institutions and imaging protocols
Wavelet-based radiomics suffers from poor standardization across imaging setups. Variations in scanner types, voxel sizes, and contrast settings alter signal characteristics, leading to inconsistent wavelet decompositions. Without standardized preprocessing or wavelet parameters, features from identical tissues may differ significantly between institutions, hindering multicenter studies and clinical translation [88, 90]. Future efforts should adopt established preprocessing standards (e.g. IBSI), report wavelet parameters explicitly, and promote inter-institutional consensus protocols. Incorporating wavelet settings into broader radiomics standardization frameworks is also recommended [88, 91].
Limitations of wavelet transforms
Wavelet transforms are prone to boundary effects due to image padding, which distorts subband values near edges—especially problematic for small ROIs, like early tumors. Additionally, feature interpretability remains limited; linking subband metrics (e.g. HH3 entropy) to biological phenomena is often unclear. Over-decomposition in low-signal-to-noise ratio (SNR) modalities (e.g. PET) may further compromise reliability [88, 90]. Future works should apply boundary-handling techniques (e.g. symmetric padding, ROI cropping), correlate wavelet features with histological or clinical outcomes to enhance interpretability, and utilize noise-robust wavelets (e.g. Coiflets) for noisy data [85, 92].
Robustness testing and validation
Without rigorous testing, wavelet-derived features may lack robustness across scanners, protocols, and perturbations [93]. All features should undergo robustness evaluation using metrics, like intraclass correlation coefficient. Unstable features should be excluded early in the pipeline to ensure statistical reliability and clinical relevance [73, 87].
Discussion
This systematic review synthesized evidence from 62 studies on the application of wavelet transforms in radiomic feature extraction from medical images, particularly CT, MRI, and PET modalities. Our analysis revealed that wavelet-based approaches consistently enhance the robustness and reproducibility of radiomic features, and partly overcome key challenges in radiomics, such as noise sensitivity, imaging artifacts, and protocol variability. By decomposing images into multiscale frequency components—low-frequency approximations for structural integrity and high-frequency details for textural nuances—wavelets enable the isolation of biologically relevant patterns that traditional preprocessing methods, like Gaussian smoothing or Fourier transforms, often overlook. This multiresolution analysis not only minimizes irrelevant variance but also preserves spatial localization, making it particularly valuable for oncology applications, including tumor grading, survival prediction, and treatment response assessment.
Key findings and trends in wavelet applications
The reviewed studies demonstrate wavelet transforms’ versatility across diverse clinical scenarios. DWT, including 2D and 3D variants, were the most prevalent (appearing in ~70% of studies), excelling in tasks, like denoising (e.g. Rician noise reduction with up to +30 dB peak signal-to-noise ratio [PSNR] gains [62]) and feature enhancement for ML classification (e.g. 98% accuracy in glioma grading using wavelet-augmented random forests [RF] [63]). Advanced variants, such as stationary wavelet transforms (SWT) and DTCWT, further improved shift-invariance and reduced aliasing artifacts, yielding superior performance in fusion tasks (e.g. CT-MRI integration with entropy gains up to 5.76 [71]) and prognostic modeling (e.g. C-index 0.62–0.68 for NSCLC survival [76]).
Figure 5 shows publication trends that indicate a steady rise in wavelet-radiomics research, from 1–2 articles annually pre-2015 to a peak of 5 in 2020, reflecting growing interest amid the radiomics boom and the COVID-19-driven demand for robust imaging analytics (e.g. lesion severity grading with AUC 0.910 [19]). Post-2020, the trajectory stabilized at 2–4 publications per year, possibly due to saturation in core applications and a shift toward integration with DL. This temporal distribution underscores wavelets’ evolution from foundational denoising tools (early 2000s) to integral components of hybrid ML pipelines, with over 80% of recent studies (2020–2025) combining them with classifiers like RF or support vector machines (SVM).
Software adoption patterns highlight MATLAB’s dominance (13 studies), likely due to its wavelet toolbox for rapid prototyping and IBSI-compliant implementations (e.g. 3.5× speedup in Haralick textures [60]). Python, with libraries, like PyRadiomics and scikit-learn (10 studies), emerged as a close second, favored for scalability and open-source ML integration (e.g. 99.2% accuracy in PDAC detection [64]). The equal share of “others” (e.g. 3D Slicer, R; 10 studies) and unspecified tools (8 studies) points to a fragmented ecosystem, emphasizing the need for standardized pipelines to ensure reproducibility. Figure 6 shows the pie chart summary based on these 36 articles, illustrating this distribution and underscoring Python’s emerging preference for open-source workflows in radiomics, balanced against MATLAB’s established clinical validation strengths. Quantitative outcomes across studies were compelling: wavelet-enhanced models achieved median AUC-ROC values of 0.85–0.98 for classification tasks (e.g. GBM vs. MET differentiation [63]) and 0.70–0.89 for prognostication (e.g. pCR prediction in breast cancer [79]). Reproducibility metrics, such as ICC >0.75, were reported in 40% of studies, with wavelet filters outperforming non-wavelet baselines by 20–50% in stability (e.g. 73% reproducible features in cardiac MRI [66]). These gains were modality-agnostic, though CT dominated (55% of studies), followed by MRI (30%) and PET (15%), aligning with CT’s prevalence in oncology workflows.
Strengths and comparative advantages
Wavelets surpass classical methods, like short-time Fourier transforms or empirical mode decomposition in most scenarios by providing scale-dependent, localized decompositions that capture heterogeneous tumor microenvironments without excessive smoothing. For instance, while Fourier methods excel in global frequency analysis, they lack spatial resolution, leading to feature dilution in noisy datasets—a limitation mitigated by wavelets’ subband thresholding (e.g. +50% SNR in multidimensional denoising [65]). Integration with ML/DL amplified these benefits: hybrid models (e.g. wavelet-CNNs) achieved 96–99% accuracies in segmentation and classification [37, 38], outperforming standalone DL by 5–10% under data scarcity.
Compared to prior reviews (Table 1), this work uniquely bridges theoretical wavelet foundations (e.g. mother wavelet selection, like Daubechies or Biorthogonal) with radiomics-specific implementations, filling a gap in modality-focused syntheses. Unlike signal-centric reviews (e.g. ECG compression [10]), we emphasized imaging applications, where wavelets’ edge-preserving properties (e.g. cavity retention in dental denoising [67]) directly translate to clinical utility.
Limitations and challenges
Despite these advances, several limitations persist. Computational intensity remains a barrier: 3D/4D decompositions demand high resources, limiting scalability for large cohorts (e.g. noted in 25% of studies [65, 73]. Parameter tuning—decomposition levels, wavelet families, and thresholding—varies widely, with suboptimal choices reducing reproducibility (e.g. rotation sensitivity in 23.7% of wavelet features [78]). Small, retrospective cohorts (median n=55–121) and single-institution data (80% of studies) raise generalizability concerns, while manual segmentations introduce observer bias [63, 64].
Heterogeneity in noise models (Gaussian vs Rician) and lack of IBSI standardization across tools further complicates comparisons, as evidenced by discrepancies in feature definitions [60]. Alternatives, like empirical mode decomposition may outperform in non-stationary signals (e.g. dynamic PET), but wavelets’ structured framework makes them more amenable to automated pipelines.
Implications for clinical practice and future directions
Wavelet-radiomics holds transformative potential for precision medicine, enabling non-invasive, quantitative phenotyping that informs personalized therapies (e.g. EGFR mutation prediction with AUC 0.709 [94]). By enhancing feature stability, it could standardize multi-institutional trials, reducing protocol-induced variances and accelerating biomarker discovery.
Future research should prioritize: 1) prospective, multicenter validations with diverse cohorts; 2) automated hyperparameter optimization via genetic algorithms or Bayesian methods to mitigate selection biases; 3) deeper DL synergies, such as wavelet-embedded CNNs for end-to-end pipelines; and 4) IBSI-compliant benchmarks for filter standardization e.g. expanding framework 4). Developing lightweight, cloud-based tools could democratize access, bridging the MATLAB-Python divide.
In conclusion, wavelet transforms represent a cornerstone for robust radiomics, offering a mathematically grounded yet practically viable pathway to overcome imaging heterogeneity. This review’s framework—encompassing workflows, challenges, and optimizations—equips researchers and clinicians to harness these tools for enhanced diagnostic and prognostic accuracy, ultimately advancing patient-centered care.
Conclusion
This systematic review confirms that wavelet transforms are a transformative approach to extracting radiomic features, leveraging their mathematical foundation in multiresolution analysis to decompose medical images into multiscale representations with exceptional accuracy. The most important findings show that wavelet-based methods, especially the DWT and advanced techniques, such as the DTCWT, enhance the detection of morphological and histological features, improve the reproducibility of features in CT, MRI, and PET modalities, and reduce the sensitivity to noise, thereby enhancing the quality and biological relevance of radiomic analyses. These capabilities have been demonstrated by their successful application in predicting complete pathological response to neoadjuvant chemotherapy in breast cancer and in stratifying hepatocellular carcinoma using MRI. However, challenges, such as parameter selection, computational complexity, and lack of standardized protocols remain significant obstacles that require robust implementation strategies and adherence to frameworks, such as the IBSI. Looking ahead, integrating wavelet-derived features with ML and DL holds promise for improving diagnostic accuracy and prognostic modeling, while multimodal approaches that combine radiomics with genomic data could provide deeper insights into disease mechanisms. Emerging applications, such as real-time intraoperative guidance and longitudinal treatment monitoring, highlight the potential of wavelet transforms for advancing precision medicine. To fully realize this potential, future efforts should prioritize interdisciplinary collaboration, validation across diverse datasets, and expansion of standardization initiatives. By addressing these gaps, wavelet-based radiomics can become a more accurate and impactful tool, bridging the gap between theoretical advances and clinical application to improve patient outcomes.
Future research should prioritize the development of hybrid approaches that integrate wavelet transforms with ML or DL to address limitations in interpretability and computational demands, the advancement of multimodal integration by combining wavelet-based radiomics with functional imaging techniques, such as PET and fMRI or with genomic and proteomic data to offer a comprehensive understanding of disease processes, and the promotion of standardization and collaboration through interdisciplinary efforts among researchers, clinicians, and industry to establish standardized protocols and open-access databases; as a result, wavelet-based radiomics emerges as a promising frontier in medical imaging, enabling advanced quantitative analysis of images, and addressing current challenges while enhancing interdisciplinary collaboration will significantly contribute to improving patient care and outcomes.
Ethical Considerations
Compliance with ethical guidelines
There were no ethical considerations to be considered in this research.
Funding
This research did not receive any grant from funding agencies in the public, commercial, or non-profit sectors.
Authors' contributions
Conceptualization and methodology: Hashem Khanbabaei and Hamid-Reza Sadoughi; Supervision: Hamid-Reza Sadoughi; Investigation, data collection and Data analysis: Hashem Khanbabaei and Sara Mohammadi; Writing the original draft, review & editing: All authors.
Conflict of interest
The authors declared no conflicts of interest.
Acknowledgments
The authors express their gratitude to the Vice-Chancellery for Research and Technology, North Khorasan University of Medical Sciences, for providing institutional support and resources that facilitated this work. They also acknowledge the support of the library services at Kerman University of Medical Sciences and Gonabad University of Medical Sciences for access to research databases.
References
- Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: The bridge between medical imaging and personalized medicine. Nature Reviews. Clinical Oncology. 2017; 14(12):749-62. [DOI:10.1038/nrclinonc.2017.141] [PMID]
- Parekh V, Jacobs MA. Radiomics: A new application from established techniques. Expert Review of Precision Medicine and Drug Development. 2016; 1(2):207-26. [DOI:10.1080/23808993.2016.1164013] [PMID]
- Wang JZ. Wavelets and imaging informatics: A review of the literature. Journal for Biomedical Informatics. 2001; 34(2):129-41. [DOI:10.1006/jbin.2001.1010] [PMID]
- Whybra P, Zwanenburg A, Andrearczyk V, Schaer R, Apte AP, Ayotte A, et al. The image biomarker standardization initiative: Standardized convolutional filters for reproducible radiomics and enhanced clinical insights. Radiology. 2024; 310(2):e231319. [DOI:10.1148/radiol.231319] [PMID]
- Dragomiretskiy K. Variational methods in signal decomposition and image processing [doctoral thesis]. Los Angeles: University of California; 2015. [Link]
- Shirazinodeh A, Noubari HA, Rabbani H, Dehnavi AM. Detection and classification of Breast Cancer in Wavelet Sub-bands of Fractal Segmented Cancerous Zones. Journal of Medical Signals and Sensors. 2015; 5(3):162-70. [DOI:10.4103/2228-7477.161486] [PMID]
- Demircioğlu A. The effect of preprocessing filters on predictive performance in radiomics. European Radiology Experimental. 2022; 6(1):40. [DOI:10.1186/s41747-022-00294-w] [PMID]
- Grobbelaar M, Phadikar S, Ghaderpour E, Struck AF, Sinha N, Ghosh R, et al. A Survey on Denoising Techniques of Electroencephalogram Signals Using Wavelet Transform. Signals. 2022; 3(3):577-86. [DOI:10.3390/signals3030035]
- Guo T, Zhang J, Lim E, López-Benítez M, Ma F, Yu L. A review of wavelet analysis and its applications: Challenges and opportunities. IEEE Access. 2022; 10:58869-903. [DOI:10.1109/ACCESS.2022.3179517]
- Manikandan MS, Dandapat S. Wavelet-based electrocardiogram signal compression methods and their performances: A prospective review. Biomedical Signal Processing and Control. 2014; 14:73-107. [DOI:10.1016/j.bspc.2014.07.002]
- Serhal H, Abdallah N, Marion JM, Chauvet P, Oueidat M, Humeau-Heurtier A. Overview on prediction, detection, and classification of atrial fibrillation using wavelets and AI on ECG. Computers in Biology and Medicine. 2022; 142:105168. [DOI:10.1016/j.compbiomed.2021.105168] [PMID]
- Shuvo SB, Alam SS, Ayman SU, Chakma A, Salvi M, Seoni S, et al. Application of wavelet transformation and artificial intelligence techniques in healthcare: A systemic review. WIREs Data Mining and Knowledge Discovery. 2025; 15(2):e70007. [DOI:10.1002/widm.70007]
- Han B. Framelets and wavelets. Cham: Birkhäuser; 2017. [DOI:10.1007/978-3-319-68530-4]
- Harpen MD. An introduction to wavelet theory and application for the radiological physicist. Medical Physics. 1998; 25(10):1985-93. [DOI:10.1118/1.598387] [PMID]
- Walnut DF. An introduction to wavelet analysis. Berlin: Springer Science & Business Media; 2013. [Link]
- Toufik B, Mokhtar N. The wavelet transform for image processing applications. Advances in Wavelet Theory and Their Applications in Engineering, Physics and Technology. 2012; 17:395-422. [Link]
- Cheng Z, Huang Y, Huang X, Wu X, Liang C, Liu Z. Effects of different wavelet filters on correlation and diagnostic performance of radiomics features. Journal of Central South University. Medical Sciences. 2019; 44(3):244-50. [DOI:10.11817/j.issn.1672-7347.2019.03.003] [PMID]
- Dumitrescu C, Raboaca MS, Felseghi RA. Methods for improving image quality for contour and textures analysis using new wavelet methods. Applied Sciences. 2021; 11(9):3895. [DOI:10.3390/app11093895]
- Jiang Z, Yin J, Han P, Chen N, Kang Q, Qiu Y, et al. Wavelet transformation can enhance computed tomography texture features: a multicenter radiomics study for grade assessment of COVID-19 pulmonary lesions. Quantitative Imaging in Medicine and Surgery. 2022; 12(10):4758-70. [DOI:10.21037/qims-22-252] [PMID]
- Garg N, Choudhry MS, Bodade RM. Alzheimer’s Disease Classification Using Wavelet-Based Image Features. Traitement du Signal. 2024; 41(4):1899. [DOI:10.18280/ts.410420]
- Mahon RN. Advanced Imaging Analysis for Predicting Tumor Response and Improving Contour Delineation Uncertainty [doctoral thesis]. Richmond: Virginia Commonwealth University; 2018. [Link]
- Chaddad A, Daniel P, Niazi T. Radiomics evaluation of histological heterogeneity using multiscale textures derived from 3D wavelet transformation of multispectral images. Frontiers in Oncology. 2018; 8:96. [DOI:10.3389/fonc.2018.00096] [PMID]
- Davnall F, Yip CS, Ljungqvist G, Selmi M, Ng F, Sanghera B, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012; 3(6):573-89. [DOI:10.1007/s13244-012-0196-6] [PMID]
- Lo SC, Li H, Freedman MT. Optimization of wavelet decomposition for image compression and feature preservation. IEEE Transactions on Medical Imaging. 2003; 22(9):1141-51. [DOI:10.1109/TMI.2003.816953] [PMID]
- Rabbani H, Vafadust M, Abolmaesumi P, Gazor S. Speckle noise reduction of medical ultrasound images in complex wavelet domain using mixture priors. IEEE Transactions on Biomedical Engineering. 2008; 55(9):2152-60. [DOI:10.1109/TBME.2008.923140] [PMID]
- Rabbani H, Nezafat R, Gazor S. Wavelet-domain medical image denoising using bivariate laplacian mixture model. IEEE Transactions on Biomedical Engineering. 2009; 56(12):2826-37. [DOI:10.1109/TBME.2009.2028876] [PMID]
- Dhawan AP. Medical image analysis. John Wiley & Sons; 2011. [DOI:10.1002/9780470918548]
- Rai R, Barton MB, Chlap P, Liney G, Brink C, Vinod S, et al. Repeatability and reproducibility of magnetic resonance imaging-based radiomic features in rectal cancer. Journal of Medical Imaging. 2022; 9(4):044005. [DOI:10.1117/1.JMI.9.4.044005] [PMID]
- Wang XH, Jiao Y, Li L. Mapping individual voxel-wise morphological connectivity using wavelet transform of voxel-based morphology. Plos One. 2018; 13(7):e0201243. [DOI:10.1371/journal.pone.0201243] [PMID]
- Wu W, Wang Y, Liu Q, Wang G, Zhang J. Wavelet-Improved Score-Based Generative Model for Medical Imaging. IEEE Transactions on Medical Imaging. 2024; 43(3):966-79. [DOI:10.1109/TMI.2023.3325824] [PMID]
- Ghalati MK, Nunes A, Ferreira H, Serranho P, Bernardes R. Texture Analysis and Its Applications in Biomedical Imaging: A Survey. IEEE Reviews in Biomedical Engineering. 2022; 15:222-46. [DOI:10.1109/RBME.2021.3115703] [PMID]
- Mingqiang Y, Kidiyo K, Joseph R. A survey of shape feature extraction techniques. Pattern Recognition. 2008; 15(7):43-90. [DOI:10.5772/6237]
- Tsai NC, Chen HW, Hsu SL. Computer-aided diagnosis for early-stage breast cancer by using Wavelet Transform. Computerized Medical Imaging and Graphics. 2011; 35(1):1-8. [DOI:10.1016/j.compmedimag.2010.08.005] [PMID]
- Wang S, Lu S, Dong Z, Yang J, Yang M, Zhang Y. Dual-Tree Complex Wavelet Transform and Twin Support Vector Machine for Pathological Brain Detection. Applied Sciences. 2016; 6(6). [DOI:10.3390/app6060169]
- Tang VH, Duong STM, Nguyen CDT, Huynh TM, Duc VT, Phan C, et al. Wavelet radiomics features from multiphase CT images for screening hepatocellular carcinoma: Analysis and comparison. The Science Reports. 2023; 13(1):19559. [DOI:10.1038/s41598-023-46695-8] [PMID]
- Lutsyk M, Gourevich K, Keidar Z. Complete pathologic response prediction by radiomics wavelets features of unenhanced ct simulation images in locally advanced rectal cancer patients after neoadjuvant chemoradiation. The Israel Medical Association Journal. 2021; 23(12):805-10. [PMID]
- Sarhan AM. Brain tumor classification in magnetic resonance images using deep learning and wavelet transform. Journal of Biomedical Science and Engineering. 2020; 13(06):102. [DOI:10.4236/jbise.2020.136010]
- Çinarer G, Emiroğlu BG, Yurttakal AH. Prediction of Glioma Grades Using Deep Learning with Wavelet Radiomic Features. Applied Sciences. 2020; 10(18):6296. [DOI:10.3390/app10186296]
- Liu G, Huang SY, Franc B, Seo Y, Mitra D. Unsupervised Learning in PET Radiomics. IEEE Nuclear Science Symposium Conference Record. 2017; 2017. [DOI:10.1109/NSSMIC.2017.8532959]
- Turkheimer FE, Aston JA, Banati RB, Riddell C, Cunningham VJ. A linear wavelet filter for parametric imaging with dynamic PET. IEEE Transactions on Medical Imaging. 2003; 22(3):289-301. [DOI:10.1109/TMI.2003.809597] [PMID]
- Li J, Mohamed SS, Salama MM, Freeman GH. Prostate tissue texture feature extraction for cancer recognition in TRUS images using wavelet decomposition. Paper presented at: International Conference Image Analysis and Recognition. 2007 August 22; Berlin, Heidelberg. [Link]
- Khalighi S, Reddy K, Midya A, Pandav KB, Madabhushi A, Abedalthagafi M. Artificial intelligence in neuro-oncology: Advances and challenges in brain tumor diagnosis, prognosis, and precision treatment. NPJ Precision Oncology. 2024; 8(1):80. [DOI:10.1038/s41698-024-00575-0] [PMID]
- Yoo Y, Lee KW, Kwon BD. Application of the 3D discrete wavelet transformation scheme to remotely sensed image classification. Korean Journal of Remote Sensing. 2007; 23(5):355-63. [Link]
- Camastra C, Pasini G, Stefano A, Russo G, Vescio B, Bini F, et al. Development and Implementation of an Innovative Framework for Automated Radiomics Analysis in Neuroimaging. Journal of Imaging. 2024; 10(4). [DOI:10.3390/jimaging10040096] [PMID]
- Prinzi F, Militello C, Conti V, Vitabile S. Impact of wavelet kernels on predictive capability of radiomic features: A case study on covid-19 chest X-ray Images. Journal of Imaging. 2023; 9(2). [DOI:10.3390/jimaging9020032] [PMID]
- Sharan V, Keshari N, Mondal T. Biomedical image denoising and compression in wavelet using MATLAB. International Journal of Innovative Science and Modern Engineering. 2014; 2319-6386. [Link]
- Jin Y, Angelini E, Laine A. Wavelets in medical image processing: Denoising, segmentation, and registration. Handbook of Biomedical Image Analysis: Volume I: Segmentation Models Part A. 2005; 305-58. [DOI:10.1007/0-306-48551-6_6]
- Misiti M, Misiti Y, Oppenheim G, Poggi JM. Wavelet toolbox. Natick: MathWorks; 1996. [Link]
- Ibanezlease L, Schroeder W, Ng L, Cates J. The ITK software guide. Clifton Park: Kitware; 2005. [Link]
- Pieper S, Halle M, Kikinis R. 3D Slicer. 2004 2nd IEEE international symposium on biomedical imaging: Nano to macro (IEEE Cat No 04EX821). IEEE. 2004. [DOI:10.1109/ISBI.2004.1398617]
- Whitcher B, Whitcher MB. Package ‘waveslim’. Waterloo: Citeseer; 2024. [Link]
- Dai Z, Chen J, Lin F, Chen Y, Fan Y, Jiang C, et al. MSD-HAM-Net: A Multi-modality Fusion Network of PET/CT Images for the Prognosis of DLBCL Patients. Artificial Neural Networks and Machine Learning - ICANN. Cham: Springer Nature Switzerland; 2024. [DOI:10.1007/978-3-031-72353-7_23]
- PyWavelets Documentation. Discrete wavelet transform (DWT) [Internet]. 2026 [Updated 2026 January 31]. Available from: [Link]
- MATLAB Help Center. denosing signal using wavedec and waverec [Internet]. 2026 [Updated 2026 January 31]. Available from: [Link]
- MATLAB Help Center. Removing noise along and within range Frequency [Internet]. 2026 [Updated 2026 January 31]. Available from: [Link]
- PyWavelets Documentation. PyWavelets - Wavelet Transforms in Python [Internet]. 2026 [Updated 2026 January 31]. Available from: [Link]
- Madero Orozco H, Vergara Villegas OO, Cruz Sánchez VG, Ochoa Domínguez H, Nandayapa Alfaro M. Automated system for lung nodules classification based on wavelet feature descriptor and support vector machine. BioMedical Engineering OnLine. 2015; 14(1):9. [DOI:10.1186/s12938-015-0003-y] [PMID]
- Hajiabadi M, Alizadeh Savareh B, Emami H, Bashiri A. Comparison of wavelet transformations to enhance convolutional neural network performance in brain tumor segmentation. BMC Medical Informatics and Decision Making. 2021; 21(1):327. [DOI:10.1186/s12911-021-01687-4] [PMID]
- Gatidis S, Hepp T, Früh M, La Fougère C, Nikolaou K, Pfannenberg C, et al. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions. Scientific Data. 2022; 9(1):601. [DOI:10.1038/s41597-022-01718-3] [PMID]
- Apte AP, Iyer A, Crispin‐Ortuzar M, Pandya R, Van Dijk LV, Spezi E, et al. Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research. Medical Physics. 2018; 45(8):3713-20. [DOI:10.1002/mp.13046] [PMID]
- Bagher-Ebadian H, Siddiqui F, Liu C, Movsas B, Chetty IJ. On the impact of smoothing and noise on robustness of CT and CBCT radiomics features for patients with head and neck cancers. Medical Physics. 2017; 44(5):1755-70. [DOI:10.1002/mp.12188] [PMID]
- Benhassine NE, Boukaache A, Boudjehem D. Medical image denoising using optimal thresholding of wavelet coefficients with selection of the best decomposition level and mother wavelet. International Journal of Imaging Systems and Technology. 2021; 31(4):1906-20. [DOI:10.1002/ima.22589]
- Bijari S, Jahanbakhshi A, Hajishafiezahramini P, Abdolmaleki P. Differentiating glioblastoma multiforme from brain metastases using multidimensional radiomics features derived from MRI and multiple machine learning models. BioMed Research International. 2022; 2022(1):2016006. [DOI:10.1155/2022/2016006] [PMID]
- Chu LC, Park S, Kawamoto S, Fouladi DF, Shayesteh S, Zinreich ES, et al. Utility of CT radiomics features in differentiation of pancreatic ductal adenocarcinoma from normal pancreatic tissue. American Journal of Roentgenology. 2019; 213(2):349-57. [DOI:10.2214/AJR.18.20901] [PMID]
- Georgieva V, Petrov P, Zlatareva D. Medical image processing based on multidimensional wavelet transforms - Advantages and trends. AIP Conference Proceedings. 2021; 2333(1). [DOI:10.1063/5.0041869]
- Jang J, Ngo LH, Mancio J, Kucukseymen S, Rodriguez J, Pierce P, et al. Reproducibility of segmentation-based myocardial radiomic features with cardiac MRI. Radiology: Cardiothoracic Imaging. 2020; 2(3):e190216. [DOI:10.1148/ryct.2020190216] [PMID]
- Kafieh R, Rabbani H, Foroohandeh M. Circular symmetric laplacian mixture model in wavelet diffusion for dental image denoising. Journal of Medical Signals and Sensors. 2012; 2(2):103-11. [DOI:10.4103/2228-7477.110447] [PMID]
- Kumar R, Gupta A, Arora HS, Pandian GN, Raman B. CGHF: A Computational Decision Support System for Glioma Classification Using Hybrid Radiomics- and Stationary Wavelet-Based Features. IEEE Access. 2020; 8:79440-58. [DOI:10.1109/ACCESS.2020.2989193]
- Larue RT, Klaassen R, Jochems A, Leijenaar RT, Hulshof MC, van Berge Henegouwen MI, et al. Pre-treatment CT radiomics to predict 3-year overall survival following chemoradiotherapy of esophageal cancer. Acta Oncologica. 2018; 57(11):1475-81. [DOI:10.1080/0284186X.2018.1486039] [PMID]
- Larue RTHM, Van De Voorde L, van Timmeren JE, Leijenaar RTH, Berbée M, Sosef MN, et al. 4DCT imaging to assess radiomics feature stability: An investigation for thoracic cancers. Radiotherapy and Oncology. 2017; 125(1):147-53. [DOI:10.1016/j.radonc.2017.07.023] [PMID]
- Moshantat M, Karamzadeh S. CT and MRI Medical Image Fusion Using Discrete Wavelet Transform. Cumhuriyet Science Journal. 2019; 40(4):939-45. [DOI:10.17776/csj.549192]
- Mukhopadhyay S, Mandal JK. Wavelet based Denoising of Medical Images Using Sub-band Adaptive Thresholding through Genetic Algorithm. Procedia Technology. 2013; 10:680-9. [DOI:10.1016/j.protcy.2013.12.410]
- Pradhan PS, King RL, Younan NH, Holcomb DW. Estimation of the Number of Decomposition Levels for a Wavelet-Based Multiresolution Multisensor Image Fusion. IEEE Transactions on Geoscience and Remote Sensing. 2006; 44(12):3674-86. [DOI:10.1109/TGRS.2006.881758]
- Procházka A, Gráfová L, Vyšata O, Caregroup N. Three-dimensional wavelet transform in multi-dimensional biomedical volume processing. Proc of the IASTED International Conference on Graphics and Virtual Reality. 2011; Cambridge.[DOI:10.2316/P.2011.741-010]
- Qiu JJ, Yin J, Ji L, Lu CY, Li K, Zhang YG, et al. Differential diagnosis of hepatocellular carcinoma and hepatic hemangioma based on maximum wavelet-coefficient statistics: Novel radiomics features from plain CT. Information Processing & Management. 2022; 59(5):103046. [DOI:10.1016/j.ipm.2022.103046]
- Soufi M, Arimura H, Nagami N. Identification of optimal mother wavelets in survival prediction of lung cancer patients using wavelet decomposition-based radiomic features. Medical Physics. 2018; 45(11):5116-28. [DOI:10.1002/mp.13202] [PMID]
- Suryanarayana G, Chandran K, Khalaf OI, Alotaibi YA, Alsufyani A, Alghamdi SA. Accurate Magnetic Resonance Image Super-Resolution Using Deep Networks and Gaussian Filtering in the Stationary Wavelet Domain. IEEE Access. 2021; 9:71406-17. [DOI:10.1109/ACCESS.2021.3077611]
- Wong LM, Ai Q, Leung HS, So TY, Hung KF, Chan YT, et al. Decoding the Rotation Effect: A Retrospective Analysis of Lesion Orientation and Its Impact on Wavelet-Based Radiomics Feature Extraction and Lung Cancer Classification. Journal of Imaging Informatics in Medicine. 2025; 1-12. [DOI:10.1007/s10278-025-01520-8]
- Zhou J, Lu J, Gao C, Zeng J, Zhou C, Lai X, et al. Predicting the response to neoadjuvant chemotherapy for breast cancer: Wavelet transforming radiomics in MRI. BMC Cancer. 2020; 20(1):100. [DOI:10.1186/s12885-020-6523-2] [PMID] []
- Kutil R. High dimensional wavelet transforms on MIMD architectures: Citeseer; 2000.
- MATLAB Help Center. Introduction to Wavelet Families [Internet]. 2026 [Updated 2026 January 31]. Available from: [Link]
- Pyradiomics. Radiomic Features [Internet]. 2026 [Updated 2026 January 31]. Available from: [Link]
- Pyradiomics. Customizing the extraction [Internet]. 2026 [Updated 2026 January 31]. Available from: [Link]
- Pyradiomics. Pipeline modules [Internet]. 2026 [Updated 2026 January 31]. Available from: [Link]
- Aerts H, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Communications. 2014; 5(1):4006. [DOI:10.1038/ncomms5006] [PMID]
- Cincotti G, Loi G, Pappalardo M. Frequency decomposition and compounding of ultrasound medical images with wavelet packets. IEEE Transactions on Medical Imaging. 2001; 20(8):764-71. [DOI:10.1109/42.938244] [PMID]
- Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, et al. Radiomics: The facts and the challenges of image analysis. European Radiology Experimental. 2018; 2(1):36. [DOI:10.1186/s41747-018-0068-z] [PMID]
- Zwanenburg A, Vallières M, Abdalah MA, Aerts H, Andrearczyk V, Apte A, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020; 295(2):328-38. [DOI:10.1148/radiol.2020191145] [PMID]
- van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research. 2017; 77(21):e104-e7. [DOI:10.1158/0008-5472.CAN-17-0339] [PMID]
- Paquier Z, Chao SL, Acquisto A, Fenton C, Guiot T, Dhont J, et al. Radiomics software comparison using digital phantom and patient data: IBSI-compliance does not guarantee concordance of feature values. Biomedical Physics & Engineering Express. 2022; 8(6). [DOI:10.1088/2057-1976/ac8e6f] [PMID]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016; 278(2):563-77. [DOI:10.1148/radiol.2015151169] [PMID]
- Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: The need for standardized methodology in tumor texture analysis. Scientific Reports. 2015; 5:11075. [DOI:10.1038/srep11075] [PMID]
- Stefano A. Challenges and limitations in applying radiomics to PET imaging: Possible opportunities and avenues for research. Computers in Biology and Medicine. 2024; 179:108827. [DOI:10.1016/j.compbiomed.2024.108827] [PMID]
- Liu Y, Kim J, Balagurunathan Y, Li Q, Garcia AL, Stringfield O, et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clinical Lung Cancer. 2016; 17(5):441-8.e6. [DOI:10.1016/j.cllc.2016.02.001] [PMID]
Type of Study: Review Article |
Subject:
● Artificial Intelligence
Received: 2025/05/18 | Accepted: 2025/10/28 | Published: 2025/12/31
Received: 2025/05/18 | Accepted: 2025/10/28 | Published: 2025/12/31
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |